Blood Res.
2019 Sep;54(3):189-197. 10.5045/br.2019.54.3.189.
Favorable long-term survival using consolidation chemotherapy without allogeneic hematopoietic cell transplantation for acute myeloid leukemia with wild-type NPM1 without FLT3-ITD
- Affiliations
-
- 1Department of Hematology/Oncology, Kyungpook National University Hospital, School of Medicine, Kyungpook National University, Daegu, Korea. jhmoon@knu.ac.kr
- 2Department of Laboratory Medicine, Kyungpook National University Hospital, School of Medicine, Kyungpook National University, Daegu, Korea.
- KMID: 2459384
- DOI: http://doi.org/10.5045/br.2019.54.3.189
Abstract
- BACKGROUND
The role of allogeneic hematopoietic cell transplantation (allo-HCT) compared with consolidation chemotherapy alone in intermediate-risk acute myeloid leukemia (AML) patients with wild-type nucleophosmin/negative or a low level of Fms related tyrosine kinase 3 internal tandem duplication (NPM1(wt)/FLT3-ITD(neg/low)) has not yet been elucidated.
METHODS
In this study, we retrospectively investigated 88 patients newly diagnosed with AML who received intensive induction chemotherapy at Kyungpook National University Hospital from March 2015 to July 2017. The selection criteria included the presence of results on genetic abnormalities including NPM1 and FLT3-ITD.
RESULTS
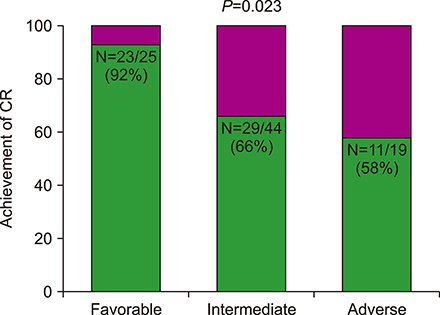
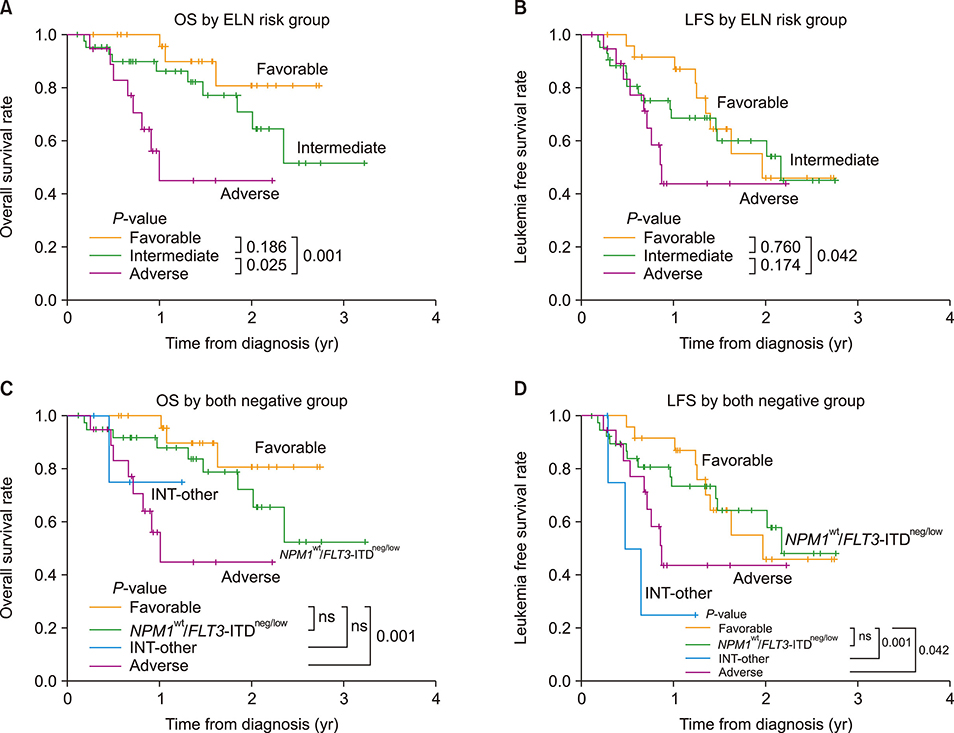
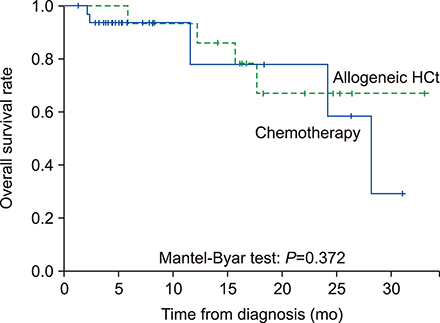
According to the European LeukemiaNet (ELN) risk classification, 25 patients (28%) were categorized as favorable, 44 (50%) as intermediate, and 19 (22%) as adverse risk. Among the intermediate-risk patients, 40 were identified as NPM1 wt/FLT3-ITDneg/low. Among the patients with NPM1(wt)/FLT3-ITD(neg/low), complete remission (CR) was achieved in 26 patients out of 40 (65%). One-year overall survival (OS) rate was 100% in the favorable-risk group and 87.9% in the NPM1(wt)/FLT3-ITD(neg/low) group (P=0.233). Among the intermediate-risk NPM1(wt)/FLT3-ITD(neg/low) patients, there was no survival benefit with allo-HCT (N=19) compared to consolidation chemotherapy (N=21; P=0.372). In the multivariate analysis, the ELN risk group [hazard ratio (HR), 6.36; P=0.019] and the achievement of CR (HR, 2.95; P=0.017) were both identified as factors affecting OS of patients with newly diagnosed AML.
CONCLUSION
Among the AML patients, intermediate-risk NPM1(wt)/FLT3-ITD(neg/low) patients and favorable-risk patients showed similar OS rates. Our results suggested that allo-HCT might have limited clinical benefit for the intermediate-risk NPM1(wt)/FLT3-ITD(neg/low) patients. Well controlled studies are needed to confirm the current results.
MeSH Terms
Figure
Reference
-
1. Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015; 373:1136–1152.
Article2. Papaemmanuil E, Dohner H, Campbell PJ. Genomic classification in acute myeloid leukemia. N Engl J Med. 2016; 375:900–901.
Article3. Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004; 350:1617–1628.
Article4. Byrd JC, Mrozek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002; 100:4325–4336.
Article5. Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004; 350:1605–1616.
Article6. Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010; 363:2424–2433.
Article7. Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013; 368:2059–2074.8. Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012; 150:264–278.
Article9. Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012; 366:1090–1098.
Article10. Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014; 506:328–333.
Article11. Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012; 366:1079–1089.12. Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016; 374:2209–2221.
Article13. Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016; 127:29–41.
Article14. Meyer SC, Levine RL. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 2014; 15:e382–e394.
Article15. Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017; 129:424–447.
Article16. Koreth J, Schlenk R, Kopecky KJ, et al. Allogeneic stem cell transplantation for acute myeloid leukemia in first complete remission: systematic review and meta-analysis of prospective clinical trials. JAMA. 2009; 301:2349–2361.
Article17. Cornelissen JJ, Blaise D. Hematopoietic stem cell transplantation for patients with AML in first complete remission. Blood. 2016; 127:62–70.
Article18. Gale RE, Green C, Allen C, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008; 111:2776–2784.
Article19. Pratcorona M, Brunet S, Nomdedeu J, et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: relevance to post-remission therapy. Blood. 2013; 121:2734–2738.
Article20. Schlenk RF, Kayser S, Bullinger L, et al. Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood. 2014; 124:3441–3449.
Article21. Heidrich K, Thiede C, Schäfer-Eckart K, et al. Allogeneic hematopoietic cell transplantation in intermediate risk acute myeloid leukemia negative for FLT3-ITD, NPM1- or biallelic CEBPA mutations. Ann Oncol. 2017; 28:2793–2798.
Article22. Ahn JS, Kim HJ, Kim YK, et al. Transplant outcomes of the triple-negative NPM1/FLT3-ITD/CEBPA mutation subgroup are equivalent to those of the favourable ELN risk group, but significantly better than the intermediate-I risk group after allogeneic transplant in normal-karyotype AML. Ann Hematol. 2016; 95:625–635.
Article23. McGowan-Jordan J, Simons A, Schmid M. ISCN 2016: an international system for human cytogenomic nomenclature (2016). Basel, Switzerland: S. Karger;2016.24. Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002; 99:4326–4335.
Article25. Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006; 107:4011–4020.
Article26. Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008; 358:1909–1918.
Article27. Pasic I, Da'na W, Lam W, et al. Influence of FLT3-ITD and NPM1 status on allogeneic hematopoietic cell transplant outcomes in patients with cytogenetically normal AML. Eur J Haematol. 2019; 102:368–374.
Article28. Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018; 378:1189–1199.
Article29. Kim T, Moon JH, Ahn JS, et al. Next-generation sequencing-based posttransplant monitoring of acute myeloid leukemia identifies patients at high risk of relapse. Blood. 2018; 132:1604–1613.
Article30. Mori A, Deola S, Xumerle L, Mijatovic V, Malerba G, Monsurro V. Next generation sequencing: new tools in immunology and hematology. Blood Res. 2013; 48:242–249.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prognostic significance of nucleophosmin mutations and FLT3 internal tandem duplication in adult patients with cytogenetically normal acute myeloid leukemia
- Prognostic significance of the FLT3 ITD mutation in patients with normal-karyotype acute myeloid leukemia in relapse
- Outcomes of pediatric acute myeloid leukemia patients with FLT3-ITD mutations in the pre-FLT3 inhibitor era
- Serial Determination of FLT3-ITD and NPM1 Mutations and Its Clinical Significance in Patients with MDS at Diagnosis and After Progression to AML with Myelodysplasia-related Changes
- FLT3 mutations in acute myeloid leukemia: a review focusing on clinically applicable drugs