Transl Clin Pharmacol.
2019 Jun;27(2):59-63. 10.12793/tcp.2019.27.2.59.
A review of computational drug repurposing
- Affiliations
-
- 1Department of Pharmacology, Yonsei University College of Medicine, Seoul 03722, Korea. kspark@yuhs.ac
- KMID: 2457572
- DOI: http://doi.org/10.12793/tcp.2019.27.2.59
Abstract
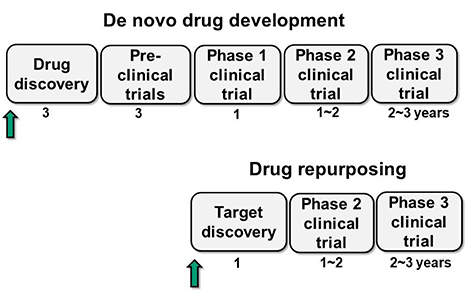
- Although sciences and technology have progressed rapidly, de novo drug development has been a costly and time-consuming process over the past decades. In view of these circumstances, "˜drug repurposing' (or "˜drug repositioning') has appeared as an alternative tool to accelerate drug development process by seeking new indications for already approved drugs rather than discovering de novo drug compounds, nowadays accounting for 30% of newly marked drugs in the U.S. In the meantime, the explosive and large-scale growth of molecular, genomic and phenotypic data of pharmacological compounds is enabling the development of new area of drug repurposing called computational drug repurposing. This review provides an overview of recent progress in the area of computational drug repurposing. First, it summarizes available repositioning strategies, followed by computational methods commonly used. Then, it describes validation techniques for repurposing studies. Finally, it concludes by discussing the remaining challenges in computational repurposing.
Keyword
Figure
Reference
-
1. Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012; 11:191–200. DOI: 10.1038/nrd3681.
Article2. Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013; 12:581–594. DOI: 10.1038/nrd4051.
Article3. Booth B, Zemmel R. Opinion/Outlook: Prospects for productivity. Nat Rev Drug Discov. 2004; 3:451–456.4. Kato S, Moulder SL, Ueno NT, Wheler JJ, Meric-Bernstam F, Kurzrock R, et al. Challenges and perspective of drug repurposing strategies in early phase clinical trials. Oncoscience. 2015; 2:576–580.
Article5. Sardana D, Zhu C, Zhang M, Gudivada RC, Yang L, Jegga AG. Drug repositioning for orphan diseases. Brief Bioinform. 2011; 12:346–356. DOI: 10.1093/bib/bbr021.
Article6. Munos BH, Chin WW. How to revive breakthrough innovation in the pharmaceutical industry. Sci Transl Med. 2011; 3:89cm16. DOI: 10.1126/scitranslmed.3002273.
Article7. Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013; 34:267–272. DOI: 10.1016/j.tips.2013.03.004.
Article8. Shim JS, Liu JO. Recent advances in drug repositioning for the discovery of new anticancer drugs. Int J Biol Sci. 2014; 10:654–663. DOI: 10.7150/ijbs.9224.
Article9. Liu Z, Fang H, Reagan K, Xu X, Mendrick DL, Slikker W Jr, et al. In silico drug repositioning: What we need to know. Drug Discov Today. 2013; 18:110–115. DOI: 10.1016/j.drudis.2012.08.005.10. Jin G, Wong ST. Toward better drug repositioning: Prioritizing and integrating existing methods into efficient pipelines. Drug Discov Today. 2014; 19:637–644. DOI: 10.1016/j.drudis.2013.11.005.
Article11. Hodos RA, Kidd BA, Shameer K, Readhead BP, Dudley JT. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip Rev Syst Biol Med. 2016; 8:186–210. DOI: 10.1002/wsbm.1337.12. Emig D, Ivliev A, Pustovalova O, Lancashire L, Bureeva S, Nikolsky Y, et al. Drug Target Prediction and Repositioning Using an Integrated Network-Based Approach. PLoS ONE. 2013; 8:e60618. DOI: 10.1371/journal.pone.0060618.
Article13. Swamidass SJ. Mining small-molecule screens to repurpose drugs. Brief Bioinform. 2011; 12:327–335. DOI: 10.1093/bib/bbr028.
Article14. Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, et al. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem. 2002; 45:2213–2221.
Article15. Jadamba E, Shin M. A Systematic Framework for Drug Repositioning from Integrated Omics and Drug Phenotype Profiles Using Pathway-Drug Network. BioMed Res Int. 2016; 2016:7147039. DOI: 10.1155/2016/7147039.
Article16. Jin G, Fu C, Zhao H, Cui K, Chang J, Wong ST. A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res. 2012; 72:33–44. DOI: 10.1158/0008-5472.CAN-11-2333.
Article17. Haeberle H, Dudley JT, Liu JT, Butte AJ, Contag CH. Identification of cell surface targets through meta-analysis of microarray data. Neoplasia. 2012; 14:666–669.
Article18. Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, et al. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005; 33:D562–D566.
Article19. Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006; 313:1929–1935.
Article20. Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011; 3:96ra76. DOI: 10.1126/scitranslmed.3002648.
Article21. Hebbring SJ. The challenges, advantages and future of phenome-wide association studies. Immunology. 2014; 141:157–165. DOI: 10.1111/imm.12195.
Article22. Luo Y, Thompson WK, Herr TM, Zeng Z, Berendsen MA, Jonnalagadda SR, et al. Natural Language Processing for EHR-Based Pharmacovigilance: A Structured Review. Drug Saf. 2017; 40:1075–1089. DOI: 10.1007/s40264-017-0558-6.
Article23. Xu H, Aldrich MC, Chen Q, Liu H, Peterson NB, Dai Q, et al. Validating drug repurposing signals using electronic health records: A case study of metformin associated with reduced cancer mortality. J Am Med Inform Assoc. 2015; 22:179–191. DOI: 10.1136/amiajnl-2014-002649.
Article24. Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: A method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011; 7:496. DOI: 10.1038/msb.2011.26.
Article25. Liu Z, Guo F, Gu J, Wang Y, Li Y, Wang D, et al. Similarity-based prediction for Anatomical Therapeutic Chemical classification of drugs by integrating multiple data sources. Bioinformatics. 2015; 31:1788–1795. DOI: 10.1093/bioinformatics/btv055.
Article26. Napolitano F, Zhao Y, Moreira VM, Tagliaferri R, Kere J, D'Amato M, et al. Drug repositioning: a machine-learning approach through data integration. J Cheminform. 2013; 5:30. DOI: 10.1186/1758-2946-5-30.
Article27. Wang Y, Chen S, Deng N, Wang Y. Drug repositioning by kernel based integration of molecular structure, molecular activity, and phenotype data. PLoS One. 2013; 8:e78518. DOI: 10.1371/journal.pone.0078518.
Article28. Menden MP, Iorio F, Garnett M, McDermott U, Benes CH, Ballester PJ, et al. Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLoS One. 2013; 8:e61318. DOI: 10.1371/journal.pone.0061318.
Article29. Lecun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015; 521:436–444.
Article30. Aliper A, Plis S, Artemov A, Ulloa A, Mamoshina P, Zhavoronkov A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol Pharm. 2016; 13:2524–2530. DOI: 10.1021/acs.molpharmaceut.6b00248.
Article31. Unterthiner T, Mayr A, Klambauer G, Hochreiter S. Toxicity Prediction using Deep Learning. Front Environ Sci. 2015; 3:10.32. Azuaje F. Drug interaction networks: An introduction to translational and clinical applications. Cardiovasc Res. 2013; 97:631–641. DOI: 10.1093/cvr/cvs289.
Article33. Iorio F, Rittman T, Ge H, Menden M, Saez-Rodriguez J. Transcriptional data: a new gateway to drug repositioning? Drug Discov Today. 2013; 18:350–357. DOI: 10.1016/j.drudis.2012.07.014.
Article34. Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G, et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol. 2012; 8:e1002503. DOI: 10.1371/journal.pcbi.1002503.
Article35. Wu C, Gudivada RC, Aronow BJ, Jegga AG. Computational drug repositioning through heterogeneous network clustering. BMC Syst Biol. 2013; 7 Suppl 5:S6. DOI: 10.1186/1752-0509-7-S5-S6.
Article36. Tari LB, Patel JH. Systematic drug repurposing through text mining. Methods Mol Biol. 2014; 1159:253–267. DOI: 10.1007/978-1-4939-0709-0_14.
Article37. Andronis C, Sharma A, Virvilis V, Deftereos S, Persidis A. Literature mining, ontologies and information visualization for drug repurposing. Brief Bioinform. 2011; 12:357–368. DOI: 10.1093/bib/bbr005.
Article38. Bisgin H, Liu Z, Fang H, Kelly R, Xu X, Tong W. A phenome-guided drug repositioning through a latent variable model. BMC Bioinformatics. 2014; 15:267.
Article39. Zhu Q, Tao C, Shen F, Chute CG. Exploring the pharmacogenomics knowledge base (PharmGKB) for repositioning breast cancer drugs by leveraging Web ontology language (OWL) and cheminformatics approaches. Pac Symp Biocomput. 2014; 172–182.
Article40. Chen B, Ding Y, Wild DJ. Assessing drug target association using semantic linked data. PLoS Comput Biol. 2012; 8:e1002574. DOI: 10.1371/journal.pcbi.1002574.
Article41. Guney E, Menche J, Vidal M, Barábasi AL. Network-based in silico drug efficacy screening. Nat Commun. 2016; 7:10331. DOI: 10.1038/ncomms10331.
Article42. Alaimo S, Giugno R, Pulvirenti A. Recommendation techniques for drug–target interaction prediction and drug repositioning. Methods Mol Biol. 2016; 1415:441–462. DOI: 10.1007/978-1-4939-3572-7_23.
Article43. Mei JP, Kwoh CK, Yang P, Li XL, Zheng J. Drug-target interaction prediction by learning from local information and neighbors. Bioinformatics. 2013; 29:238–245. DOI: 10.1093/bioinformatics/bts670.
Article44. Khatri P, Roedder S, Kimura N, De Vusser K, Morgan AA, Gong Y, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013; 210:2205–2221. DOI: 10.1084/jem.20122709.
Article45. Li YY, Jones SJ. Drug repositioning for personalized medicine. Genome Med. 2012; 4:27. DOI: 10.1186/gm326.
Article46. Xu Y, Dai Z, Chen F, Gao S, Pei J, Lai L. Deep learning for drug-induced liver injury. J Chem Inf Model. 2015; 55:2085–2093. DOI: 10.1021/acs.jcim.5b00238.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The use of real-world data in drug repurposing
- Anti-inflammatory Strategies for Schizophrenia: A Review of Evidence for Therapeutic Applications and Drug Repurposing
- Computational Challenges for Integrative Genomics
- Computational modeling of atrial fibrillation
- Computational Neuroscience Approach to Psychiatry: A Review on Theory-driven Approaches