Investig Clin Urol.
2016 Jul;57(4):286-297. 10.4111/icu.2016.57.4.286.
Gene expression profile comparison in the penile tissue of diabetes and cavernous nerve injury-induced erectile dysfunction rat model
- Affiliations
-
- 1Department of Urology, Gyeongsang National University Changwon Hospital, Gyeongsang National University School of Medicine, Changwon, Korea.
- 2Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, UT, USA.
- 3Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Institute of Human-Environment Interface Biology, Seoul National University, Seoul, Korea.
- 4Department of Physiology and Biophysics, Seoul National University College of Medicine, Seoul, Korea.
- 5Department of Urology, Samsung Medical Center, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Seoul, Korea. drswlee@skku.edu
- 6Department of Urology, Chonbuk National University Medical School, Institute for Medical Sciences, Chonbuk National University, Research Institute and Clinical Trial Center of Medical Device of Chonbuk National University Hospital, Jeonju, Korea.
- KMID: 2426507
- DOI: http://doi.org/10.4111/icu.2016.57.4.286
Abstract
- PURPOSE
To investigate the effects of cavernous nerve injury (CNI) on gene expression profiles in the cavernosal tissue of a CNI-induced erectile dysfunction (ED) model and to provide a basis for future investigations to discover potential target genes for ED treatment.
MATERIALS AND METHODS
Young adult rats were divided randomly into 2 groups: sham operation and bilateral CN resection. At 12 weeks after CNI we measured erectile responses and performed microarray experiments and gene set enrichment analysis to reveal gene signatures that were enriched in the CNI-induced ED model. Alterations in gene signatures were compared with those in the diabetes-induced ED model. The diabetic-induced ED data is taken from GSE2457.
RESULTS
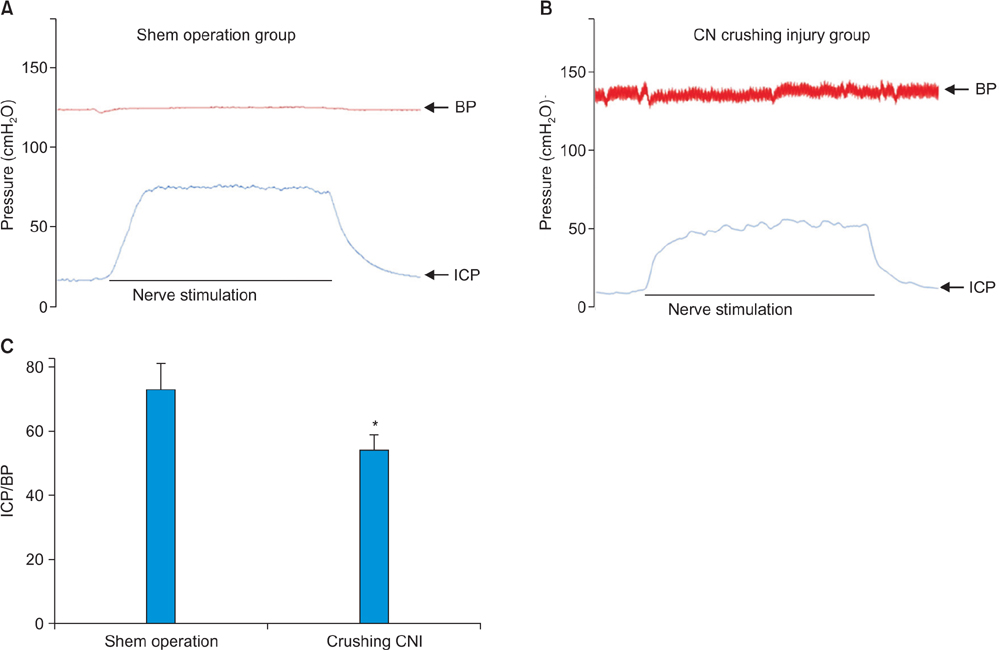
The mean ratio of intracavernosal pressure/blood pressure for the CNI group (0.54±0.4 cmHâ‚‚O) was significantly lower than that in the sham operation group (0.73±0.8 cmHâ‚‚O, p<0.05). Supervised and unsupervised clustering analysis showed that the diabetes- and CNI-induced ED cavernous tissues had different gene expression profiles from normal cavernous tissues. We identified 46 genes that were upregulated and 77 genes that were downregulated in both the CNI- and diabetes-induced ED models.
CONCLUSIONS
Our genome-wide and computational studies provide the groundwork for understanding complex mechanisms and molecular signature changes in ED.
MeSH Terms
-
Animals
Cluster Analysis
Diabetes Mellitus, Experimental/complications/*genetics/metabolism
Down-Regulation/physiology
Erectile Dysfunction/etiology/*genetics/metabolism
Gene Expression Profiling/methods
Genome-Wide Association Study
Male
Penile Erection/genetics/physiology
Penis/innervation/*metabolism
Peripheral Nerve Injuries/complications/*genetics/metabolism
*Transcriptome
Up-Regulation/physiology
Figure
Reference
-
1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010; 60:277–300.2. Noldus J, Michl U, Graefen M, Haese A, Hammerer P, Huland H. Patient-reported sexual function after nerve-sparing radical retropubic prostatectomy. Eur Urol. 2002; 42:118–124.3. Sanda MG, Dunn RL, Michalski J, Sandler HM, Northouse L, Hembroff L, et al. Quality of life and satisfaction with outcome among prostate-cancer survivors. N Engl J Med. 2008; 358:1250–1261.4. User HM, Hairston JH, Zelner DJ, McKenna KE, McVary KT. Penile weight and cell subtype specific changes in a post-radical prostatectomy model of erectile dysfunction. J Urol. 2003; 169:1175–1179.5. Mullerad M, Donohue JF, Li PS, Scardino PT, Mulhall JP. Functional sequelae of cavernous nerve injury in the rat: is there model dependency. J Sex Med. 2006; 31:77–83.6. Yamashita S, Kato R, Kobayashi K, Hisasue S, Arai Y, Tsukamoto T. Nerve injury-related erectile dysfunction following nerve-sparing radical prostatectomy: a novel experimental dissection model. Int J Urol. 2009; 16:905–911.7. McVary KT, Razzaq A, Lee C, Venegas MF, Rademaker A, McKenna KE. Growth of the rat prostate gland is facilitated by the autonomic nervous system. Biol Reprod. 1994; 51:99–107.8. Rehman J, Chenven E, Brink P, Peterson B, Walcott B, Wen YP, et al. Diminished neurogenic but not pharmacological erections in the 2- to 3-month experimentally diabetic F-344 rat. Am J Physiol. 1997; 272(4 Pt 2):H1960–H1971.9. Alers JC, Rochat J, Krijtenburg PJ, van Dekken H, Raap AK, Rosenberg C. Universal linkage system: an improved method for labeling archival DNA for comparative genomic hybridization. Genes Chromosomes Cancer. 1999; 25:301–305.10. Sullivan CJ, Teal TH, Luttrell IP, Tran KB, Peters MA, Wessells H. Microarray analysis reveals novel gene expression changes associated with erectile dysfunction in diabetic rats. Physiol Genomics. 2005; 23:192–205.11. Eijssen LM, Jaillard M, Adriaens ME, Gaj S, de Groot PJ, Muller M, et al. User-friendly solutions for microarray quality control and pre-processing on ArrayAnalysis.org. Nucleic Acids Res. 2013; 41(Web Server issue):W71–W76.12. Lee S, Chun JN, Kim SH, So I, Jeon JH. Icilin inhibits E2F1-mediated cell cycle regulatory programs in prostate cancer. Biochem Biophys Res Commun. 2013; 441:1005–1010.13. Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012; 100:337–344.14. Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005; 33:e175.15. Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor. 2009; 11:10–18.16. Cao LJ, Keerthi SS, Ong CJ, Zhang JQ, Periyathamby U, Fu XJ, et al. Parallel sequential minimal optimization for the training of support vector machines. IEEE Trans Neural Netw. 2006; 17:1039–1049.17. In : Kohavi R, editor. A study of cross-validation and bootstrap for accuracy estimation and model selection. IJCAI'95 Proceedings of the 14th international joint conference on Artificial intelligence; San Francisco (CA): Morgan Kaufmann Publishers Inc.;1995. p. 1137–1143.18. Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol. 2005; 3:185–205.19. Yu L, Liu H. Efficient feature selection via analysis of relevance and redundancy. J Mach Learn Res. 2004; 5:1205–1224.20. Hubble J, Demeter J, Jin H, Mao M, Nitzberg M, Reddy TB, et al. Implementation of GenePattern within the Stanford Microarray Database. Nucleic Acids Res. 2009; 37(Database issue):D898–D901.21. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–15550.22. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001; 98:5116–5121.23. Leungwattanakij S, Bivalacqua TJ, Usta MF, Yang DY, Hyun JS, Champion HC, et al. Cavernous neurotomy causes hypoxia and fibrosis in rat corpus cavernosum. J Androl. 2003; 24:239–245.24. Hyun JS. Prostate cancer and sexual function. World J Mens Health. 2012; 30:99–107.25. Hipp JD, Davies KP, Tar M, Valcic M, Knoll A, Melman A, et al. Using gene chips to identify organ-specific, smooth muscle responses to experimental diabetes: potential applications to urological diseases. BJU Int. 2007; 99:418–430.26. Musicki B, Liu T, Lagoda GA, Strong TD, Sezen SF, Johnson JM, et al. Hypercholesterolemia-induced erectile dysfunction: endothelial nitric oxide synthase (eNOS) uncoupling in the mouse penis by NAD(P)H oxidase. J Sex Med. 2010; 7:3023–3032.27. Hu C, Dong YY, Dong YH, Cui JF, Dai JC. Identification of oxidative stress-induced gene expression profiles in cavernosal endothelial cells. Mol Med Rep. 2015; 11:2781–2788.28. Kakoki M, McGarrah RW, Kim HS, Smithies O. Bradykinin B1 and B2 receptors both have protective roles in renal ischemia/ reperfusion injury. Proc Natl Acad Sci U S A. 2007; 104:7576–7581.29. Calenda G, Strong TD, Pavlovich CP, Schaeffer EM, Burnett AL, Yu W, et al. Whole genome microarray of the major pelvic ganglion after cavernous nerve injury: new insights into molecular profile changes after nerve injury. BJU Int. 2012; 109:1552–1564.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The neural mechanism of apomorphine-induced erection: an experimental study by the comparison with electrostimulation-induced erection of the rat model
- Differential Expression of Nerve Injury-Induced Protein 1 (Ninjurin 1) in In Vivo and In Vitro Models for Diabetic Erectile Dysfunction
- Argonaute 2 restored erectile function and corpus cavernosum mitochondrial function by reducing apoptosis in a mouse model of cavernous nerve injury
- Photobiomodulation as a Potential Therapy for Erectile Function: A Preclinical Study in a Cavernous Nerve Injury Model
- Relaxin-2 Prevents Erectile Dysfunction by Cavernous Nerve, Endothelial and Histopathological Protection Effects in Rats with Bilateral Cavernous Nerve Injury