Ann Lab Med.
2017 Nov;37(6):536-539. 10.3343/alm.2017.37.6.536.
Identification of the PROM1 Mutation p.R373C in a Korean Patient With Autosomal Dominant Stargardt-like Macular Dystrophy
- Affiliations
-
- 1Department of Ophthalmology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. sangjinkim@skku.edu
- 2Samsung Genome Institute, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 3Department of Health Sciences and Technology, SAIHST, Sungkyunkwan University, Seoul, Korea.
- 4Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 5Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 6Department of Neurology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 2425997
- DOI: http://doi.org/10.3343/alm.2017.37.6.536
Abstract
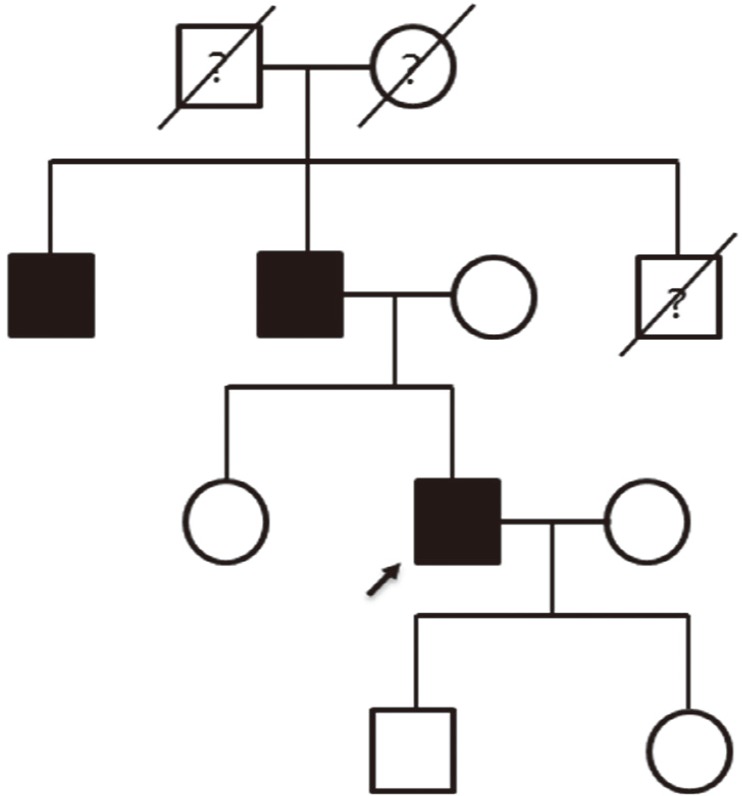
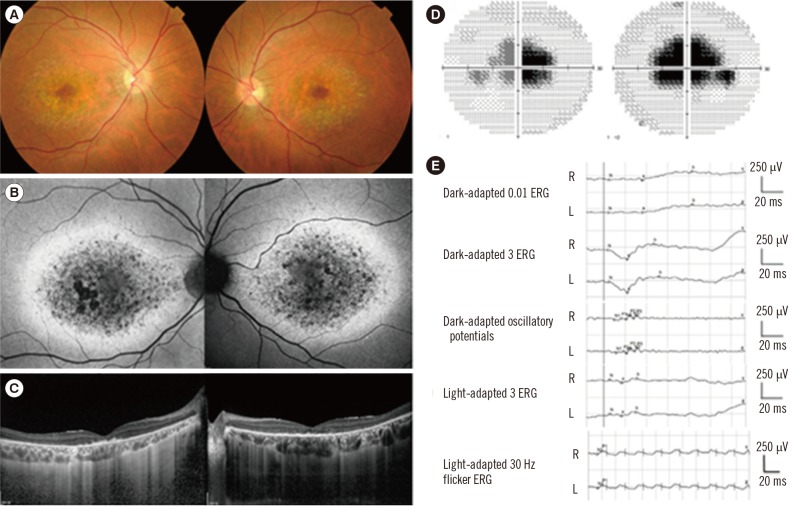
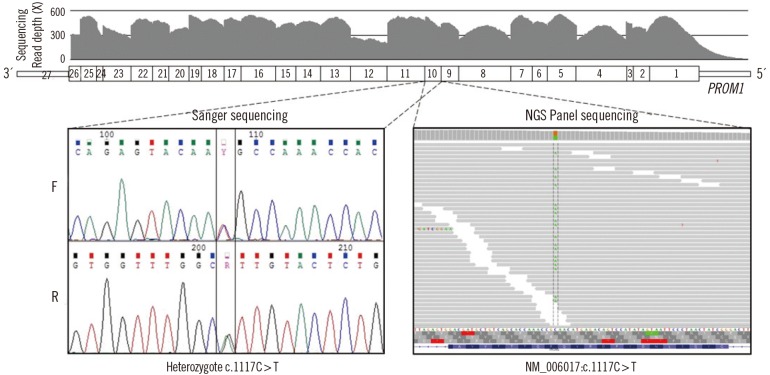
- Stargardt-like macular dystrophy 4 (STGD4) is a rare macular dystrophy characterized by bull's eye atrophy of the macula and the underlying retinal pigment epithelium. Patients with STGD4 show decreased central vision, which often progresses to severe vision loss. The PROM1 gene encodes prominin-1, which is a 5-transmembrane glycoprotein also known as CD133 and is involved in photoreceptor disk morphogenesis. PROM1 mutations have been identified as genetic causes for STGD4 and other retinal degenerations such as retinitis pigmentosa. We report a case of STGD4 with a PROM1 p.R373C mutation in a Korean patient. Ophthalmic examinations of a 38-yr old man complaining of decreased visual acuity revealed bilateral atrophic macular lesions consistent with STGD4. Targeted exome sequencing of known inherited retinal degeneration genes revealed a heterozygous missense mutation c.1117C>T (p.R373C) of PROM1, which was confirmed by Sanger sequencing. To the best of our knowledge, this is the first case of a PROM1 mutation causing STGD4 in Koreans.
MeSH Terms
Figure
Reference
-
1. Kniazeva M, Chiang MF, Morgan B, Anduze AL, Zack DJ, Han M, et al. A new locus for autosomal dominant stargardt-like disease maps to chromosome 4. Am J Hum Genet. 1999; 64:1394–1399. PMID: 10205271.2. Yang Z, Chen Y, Lillo C, Chien J, Yu Z, Michaelides M, et al. Mutant prominin 1 found in patients with macular degeneration disrupts photo-receptor disk morphogenesis in mice. J Clin Invest. 2008; 118:2908–2916. PMID: 18654668.3. Michaelides M, Gaillard MC, Escher P, Tiab L, Bedell M, Borruat FX, et al. The PROM1 mutation p.R373C causes an autosomal dominant bull’s eye maculopathy associated with rod, rod-cone, and macular dystrophy. Invest Ophthalmol Vis Sci. 2010; 51:4771–4780. PMID: 20393116.4. Palejwala NV, Gale MJ, Clark RF, Schlechter C, Weleber RG, Pennesi ME. Insights into autosomal dominant stargardt-like macular dystrophy through multimodality diagnostic imaging. Retina. 2016; 36:119–130. PMID: 26110599.5. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. PMID: 19451168.6. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. PMID: 19505943.7. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. PMID: 20644199.8. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164. PMID: 20601685.9. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–291. PMID: 27535533.10. Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003; 31:3812–3814. PMID: 12824425.11. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013; Chapter 7:Unit7.20.12. Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010; 6:e1001025. PMID: 21152010.13. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014; 42:D980–D985. PMID: 24234437.14. Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997; 15:236–246. PMID: 9054934.15. Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001; 27:89–93. PMID: 11138005.16. Michaelides M, Johnson S, Poulson A, Bradshaw K, Bellmann C, Hunt DM, et al. An autosomal dominant bull's-eye macular dystrophy (MCDR2) that maps to the short arm of chromosome 4. Invest Ophthalmol Vis Sci. 2003; 44:1657–1662. PMID: 12657606.17. Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997; 90:5002–5012. PMID: 9389720.18. Kempermann G, Chesler EJ, Lu L, Williams RW, Gage FH. Natural variation and genetic covariance in adult hippocampal neurogenesis. Proc Natl Acad Sci U S A. 2006; 103:780–785. PMID: 16407118.19. Arrigoni FI, Matarin M, Thompson PJ, Michaelides M, McClements ME, Redmond E, et al. Extended extraocular phenotype of PROM1 mutation in kindreds with known autosomal dominant macular dystrophy. Eur J Hum Genet. 2011; 19:131–137. PMID: 20859302.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Stargardt`s Macular Dystrophy
- Electroretinogrophic Finding of the Stargardt's Ddosease
- Genetic Mutation Profiles in Korean Patients with Inherited Retinal Diseases
- Cases of Macular Corneal Dystrophy with a Family History
- A Korean Family with an Early-Onset Autosomal Dominant Macular Dystrophy Resembling North Carolina Macular Dystrophy