J Vet Sci.
2017 Jun;18(2):229-236. 10.4142/jvs.2017.18.2.229.
Antibiotic resistance patterns and genetic relatedness of Enterococcus faecalis and Enterococcus faecium isolated from military working dogs in Korea
- Affiliations
-
- 1BK21 PLUS Program for Creative Veterinary Science Research, College of Veterinary Medicine and Research Institute for Veterinary Science, Seoul National University, Seoul 08826, Korea. chose@snu.ac.kr
- KMID: 2412576
- DOI: http://doi.org/10.4142/jvs.2017.18.2.229
Abstract
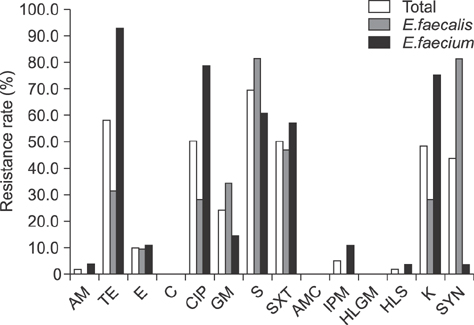
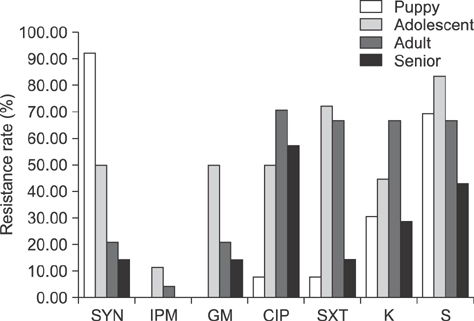
- Enterococcus spp. are normally present in the gastrointestinal tracts of animals and humans, but can cause opportunistic infections that can be transmitted to other animals or humans with integrated antibiotic resistance. To investigate if this is a potential risk in military working dogs (MWDs), we analyzed antibiotic resistance patterns and genetic relatedness of Enterococcus spp. isolated from fecal samples of MWDs of four different age groups. Isolation rates of Enterococcus spp., Enterococcus (E.) faecalis, and E. faecium, were 87.7% (57/65), 59.6% (34/57), and 56.1% (32/57), respectively, as determined by bacterial culture and multiplex PCR. The isolation rate of E. faecalis gradually decreased with age (puppy, 100%; adolescent, 91.7%; adult, 36.4%; and senior, 14.3%). Rates of resistance to the antibiotics ciprofloxacin, gentamicin, streptomycin, sulfamethoxazole/trimethoprim, imipenem, and kanamycin among Enterococcus spp. increased in adolescents and adults and decreased in senior dogs, with some isolates having three different antibiotic resistance patterns. There were indistinguishable pulsed-field gel electrophoresis patterns among the age groups. The results suggest that Enterococcus is horizontally transferred, regardless of age. As such, periodic surveillance studies should be undertaken to monitor changes in antibiotic resistance, which may necessitate modification of antibiotic regimens to manage antibiotic resistance transmission.
Keyword
MeSH Terms
-
Animals
Anti-Bacterial Agents/*therapeutic use
Dog Diseases/drug therapy/*microbiology
Dogs
Drug Resistance, Bacterial/genetics
Electrophoresis, Gel, Pulsed-Field/veterinary
Enterococcus faecalis/*drug effects/genetics
Enterococcus faecium/*drug effects/genetics
Female
Gram-Positive Bacterial Infections/drug therapy/microbiology/*veterinary
Male
Microbial Sensitivity Tests/veterinary
Military Personnel
Multiplex Polymerase Chain Reaction/veterinary
Republic of Korea
Anti-Bacterial Agents
Figure
Reference
-
1. Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012; 10:266–278.
Article2. Bortolaia V, Guardabassi L. Zoonotic transmission of antimicrobial resistant enterococci: a threat to public health or an overemphasised risk?. In : Sing A, editor. Zoonoses—Infections Affecting Humans and Animals. Focus on Public Health Aspects. New York: Springer;2015. p. 407–431.3. Buma R, Maeda T, Kamei M, Kourai H. Pathogenic bacteria carried by companion animals and their susceptibility to antibacterial agents. Biocontrol Sci. 2006; 11:1–9.
Article4. Butaye P, Devriese LA, Haesebrouck F. Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animals. Antimicrob Agents Chemother. 2001; 45:1374–1378.
Article5. Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. Enterococci in the environment. Microbiol Mol Biol Rev. 2012; 76:685–706.
Article6. Centers for Disease Control and Prevention (US). Standard operating procedure for PulseNet PFGE of Listeria monocytogenes. Atlanta: 2013.7. Chung YS, Kwon KH, Shin S, Kim JH, Park YH, Yoon JW. Characterization of veterinary hospital-associated isolates of Enterococcus species in Korea. J Microbiol Biotechnol. 2014; 24:386–393.
Article8. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI document M100-S24. Wayne: Clinical and Laboratory Standards Institute;2014.9. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. CLSI document M100-S23. Wayne: Clinical and Laboratory Standards Institute;2013.10. Damborg P, Sørensen AH, Guardabassi L. Monitoring of antimicrobial resistance in healthy dogs: first report of canine ampicillin-resistant Enterococcus faecium clonal complex 17. Vet Microbiol. 2008; 132:190–196.
Article11. Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995; 33:24–27.
Article12. Ghosh A, Dowd SE, Zurek L. Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS One. 2011; 6:e22451.
Article13. Ghosh A, Kukanich K, Brown CE, Zurek L. Resident cats in small animal veterinary hospitals carry multi-drug resistant enterococci and are likely involved in cross-contamination of the hospital environment. Front Microbiol. 2012; 3:62.
Article14. Guardabassi L, Schwarz S, Lloyd DH. Pet animals as reservoirs of antimicrobial-resistant bacteria. J Antimicrob Chemother. 2004; 54:321–332.15. Hammerum AM, Lester CH, Heuer OE. Antimicrobial-resistant enterococci in animals and meat: a human health hazard? Foodborne Pathog Dis. 2010; 7:1137–1146.
Article16. Jackson CR, Fedorka-Cray PJ, Barrett JB. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J Clin Microbiol. 2004; 42:3558–3565.
Article17. Jackson CR, Fedorka-Cray PJ, Davis JA, Barrett JB, Brousse JH, Gustafson J, Kucher M. Mechanisms of antimicrobial resistance and genetic relatedness among enterococci isolated from dogs and cats in the United States. J Appl Microbiol. 2010; 108:2171–2179.
Article18. Jackson CR, Fedorka-Cray PJ, Davis JA, Barrett JB, Frye JG. Prevalence, species distribution and antimicrobial resistance of enterococci isolated from dogs and cats in the United States. J Appl Microbiol. 2009; 107:1269–1278.
Article19. Kataoka Y, Ito C, Kawashima A, Ishii M, Yamashiro S, Harada K, Ochi H, Sawada T. Identification and antimicrobial susceptibility of enterococci isolated from dogs and cats subjected to differing antibiotic pressures. J Vet Med Sci. 2013; 75:749–753.
Article20. Kataoka Y, Umino Y, Ochi H, Harada K, Sawada T. Antimicrobial susceptibility of enterococcal species isolated from antibiotic-treated dogs and cats. J Vet Med Sci. 2014; 76:1399–1402.
Article21. Kristic CJ, Rice LB, Arias CA. Enterococcal infectiontreatment and antibiotic resistance. In : Gilmore MS, Clewell BD, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary;2014. p. 87–134.22. Levin BR, Lipsitch M, Perrot V, Schrag S, Antia R, Simonsen L, Walker NM, Stewart FM. The population genetics of antibiotic resistance. Clin Infect Dis. 1997; 24:Suppl 1. S9–S16.
Article23. Lindberg E, Adlerberth I, Hesselmar B, Saalman R, Strannegård IL, Åberg N, Wold AE. High rate of transfer of Staphylococcus aureus from parental skin to infant gut flora. J Clin Microbiol. 2004; 42:530–534.
Article24. Lloyd DH. Reservoirs of antimicrobial resistance in pet animals. Clin Infect Dis. 2007; 245:Suppl 2. S148–S152.
Article25. Moellering RC Jr. Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992; 14:1173–1176.26. Park JH. A surveillance study of antibiotic resistance of Enterococcus spp. isolated from cattle in Busan area. Annu Rep Busan Metrop City Inst Health Environ. 2013; 23:290–300.27. Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004; 12:412–416.
Article28. Van den Bogaard AE, Stobberingh EE. Epidemiology of resistance to antibiotics. Links between animals and humans. Int J Antimicrob Agents. 2000; 14:327–335.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correlation between Infective Factors and Antibiotic Resistance in Enterococci Clinical Isolates in West of Iran
- Susceptibility of Fosfomycin against Vancomycin Resistant Enterococci
- Distributions of Listeria spp., Bacillus spp., Enterococcus spp., Staphylococcus spp., and Coliforms Isolated from Agricultural Herb Products from the Market
- Characterization of Vancomycin-Resistant Enterococci Isolated from Stools and Their Acquisition of Vancomycin Resistance
- Risk Factors for Acute Cholangitis Caused by Enterococcus faecalis and Enterococcus faecium