J Breast Cancer.
2017 Dec;20(4):393-399. 10.4048/jbc.2017.20.4.393.
Optimization of RNA Extraction from Formalin-Fixed Paraffin-Embedded Blocks for Targeted Next-Generation Sequencing
- Affiliations
-
- 1Department of Pathology, Korea University Guro Hospital, Seoul, Korea. idea1@hanmail.net
- KMID: 2398210
- DOI: http://doi.org/10.4048/jbc.2017.20.4.393
Abstract
- PURPOSE
Breast cancer has a high prevalence in Korea. To achieve personalized therapy for breast cancer, long-term follow-up specimens are needed for next-generation sequencing (NGS) and multigene analysis. Formalin-fixed paraffin-embedded (FFPE) samples are easier to store than fresh frozen (FF) samples. The objective of this study was to optimize RNA extraction from FFPE blocks for NGS.
METHODS
RNA quality from FF and FFPE tissues (n=5), expected RNA amount per unit area, the relationship between archiving time and quantity/quality of FFPE-extracted RNA (n=14), differences in quantitative real-time polymerase chain reaction (qRT-PCR) and NGS results, and comparisons of both techniques with tissue processing at different institutions (n=96) were determined in this study.
RESULTS
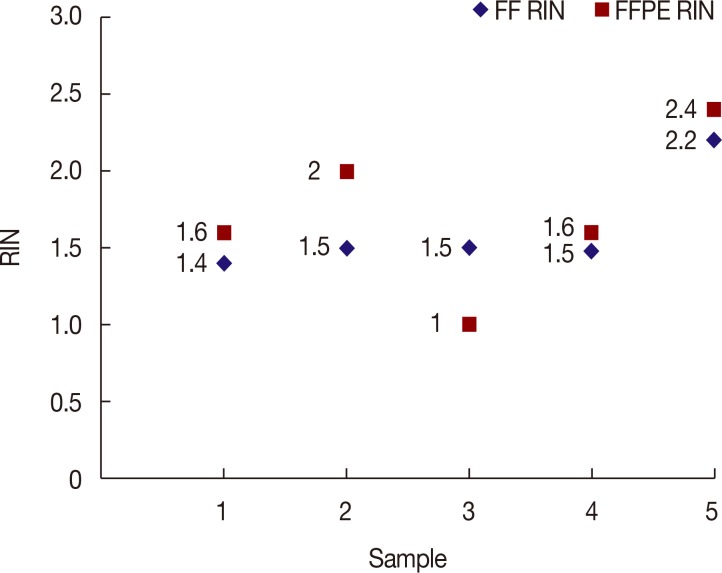
The quality of RNA did not show any statistically significant difference between paired FF and FFPE specimens (p=0.49). Analysis of tumor cellularity gave an expected RNA amount of 33.25 ng/mm2. Archiving time affected RNA quality, showing a negative correlation with RNA integrity number and a positive correlation with threshold cycle. However, RNA from samples as old as 10 years showed a 100% success rate in qRT-PCR using short primers, showing that the effect of archiving time can be overcome by proper experiment design. NGS showed a higher success rate than qRT-PCR. Specimens from institution B (n=46), which were often stored in a refrigerator for more than 6 hours and fixed without slicing, showed lower success rates and worse results than specimens from the other institutes.
CONCLUSION
Archived FFPE tissues can be used to extract RNA for NGS if they are properly processed before fixation. The expected amount of RNA per unit size calculated in this study will be useful for other researchers.
Keyword
MeSH Terms
Figure
Reference
-
1. Jung KW, Won YJ, Kong HJ, Oh CM, Lee DH, Lee JS. Prediction of cancer incidence and mortality in Korea, 2014. Cancer Res Treat. 2014; 46:124–130. PMID: 24851103.
Article2. Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014; 9:e98187. PMID: 24878701.
Article3. Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive: gene expression in paraffin-embedded tissue. J Pathol. 2001; 195:66–71. PMID: 11568892.4. Nechifor-Boilă AC, Loghin A, Vacariu V, Halaţiu VB, Borda A. The storage period of the formalin-fixed paraffin-embedded tumor blocks does not influence the concentration and purity of the isolated DNA in a series of 83 renal and thyroid carcinomas. Rom J Morphol Embryol. 2015; 56(2 Suppl):759–763. PMID: 26429169.5. Lee JE, Kim JH, Hong EJ, Yoo HS, Nam HY, Park O. National Biobank of Korea: quality control programs of collected-human biospecimens. Osong Public Health Res Perspect. 2012; 3:185–189. PMID: 24159512.
Article6. Scolnick JA, Dimon M, Wang IC, Huelga SC, Amorese DA. An efficient method for identifying gene fusions by targeted RNA sequencing from fresh frozen and FFPE samples. PLoS One. 2015; 10:e0128916. PMID: 26132974.
Article7. Ribeiro-Silva A, Zhang H, Jeffrey SS. RNA extraction from ten year old formalin-fixed paraffin-embedded breast cancer samples: a comparison of column purification and magnetic bead-based technologies. BMC Mol Biol. 2007; 8:118. PMID: 18154675.
Article8. Rupp GM, Locker J. Purification and analysis of RNA from paraffin-embedded tissues. Biotechniques. 1988; 6:56–60. PMID: 2483655.9. Tsoi DT, Inoue M, Kelly CM, Verma S, Pritchard KI. Cost-effectiveness analysis of recurrence score-guided treatment using a 21-gene assay in early breast cancer. Oncologist. 2010; 15:457–465. PMID: 20421264.
Article10. Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004; 351:2817–2826. PMID: 15591335.
Article11. Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics. 2015; 8:54. PMID: 26297356.
Article12. Prat A, Bianchini G, Thomas M, Belousov A, Cheang MC, Koehler A, et al. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH study. Clin Cancer Res. 2014; 20:511–521. PMID: 24443618.
Article13. Chung JY, Braunschweig T, Hewitt SM. Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn Mol Pathol. 2006; 15:229–236. PMID: 17122651.
Article14. Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006; 27:126–139. PMID: 16469371.
Article15. Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006; 7:3. PMID: 16448564.
Article16. Guo Y, Li CI, Ye F, Shyr Y. Evaluation of read count based RNAseq analysis methods. BMC Genomics. 2013; 14(Suppl 8):S2.
Article17. Rosen PP, Menendez-Botet CJ, Nisselbaum JS, Urban JA, Miké V, Fracchia A, et al. Pathological review of breast lesions analyzed for estrogen receptor protein. Cancer Res. 1975; 35(11 Pt 1):3187–3194. PMID: 171066.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparison of the DNA Preservation in Neutral-Buffered Formalin Fixed Paraffin-Embedded Tissue and in Non-Buffered Formalin Fixed Paraffin-Embedded Tissue
- Morule-like features in pulmonary adenocarcinoma associated with epidermal growth factor receptor mutations: two case reports with targeted next-generation sequencing analysis
- Telomerase Activity in Gastric Adenocarcinomas: Frozen Tissues Versus Methacarn-fixed Paraffin-embedded Tissues
- Multistaining Optimization for Epstein-Barr Virus–Encoded RNA In Situ Hybridization and Immunohistochemistry of Formalin-Fixed Paraffin-Embedded Tissues Using an Automated Immunostainer
- Flow Cytometric DNA Content Analysis in Breast Cancer Comparison study of fresh and paraffin-embedded tissues