Cancer Res Treat.
2017 Oct;49(4):1012-1021. 10.4143/crt.2016.433.
Identification of a Novel BRCA1 Pathogenic Mutation in Korean Patients Following Reclassification of BRCA1 and BRCA2 Variants According to the ACMG Standards and Guidelines Using Relevant Ethnic Controls
- Affiliations
-
- 1Hereditary Cancer Clinic, Cancer Prevention Center, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea. LEE.ST@yuhs.ac
- 2Department of Medicine, The Graduate School of Yonsei University, Yonsei University College of Medicine, Seoul, Korea.
- 3Department of Obstetrics and Gynecology, Institute of Women's Life Medical Science, Women's Cancer Clinic, Yonsei University College of Medicine, Seoul, Korea.
- 4Department of Surgery, Yonsei University College of Medicine, Seoul, Korea.
- 5Department of Pediatrics, Yonsei University College of Medicine, Seoul, Korea.
- 6Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea.
- 7Division of Gastroenterology, Department of Internal Medicine, Yonsei University College of Medicine, Seoul, Korea.
- KMID: 2394820
- DOI: http://doi.org/10.4143/crt.2016.433
Abstract
- PURPOSE
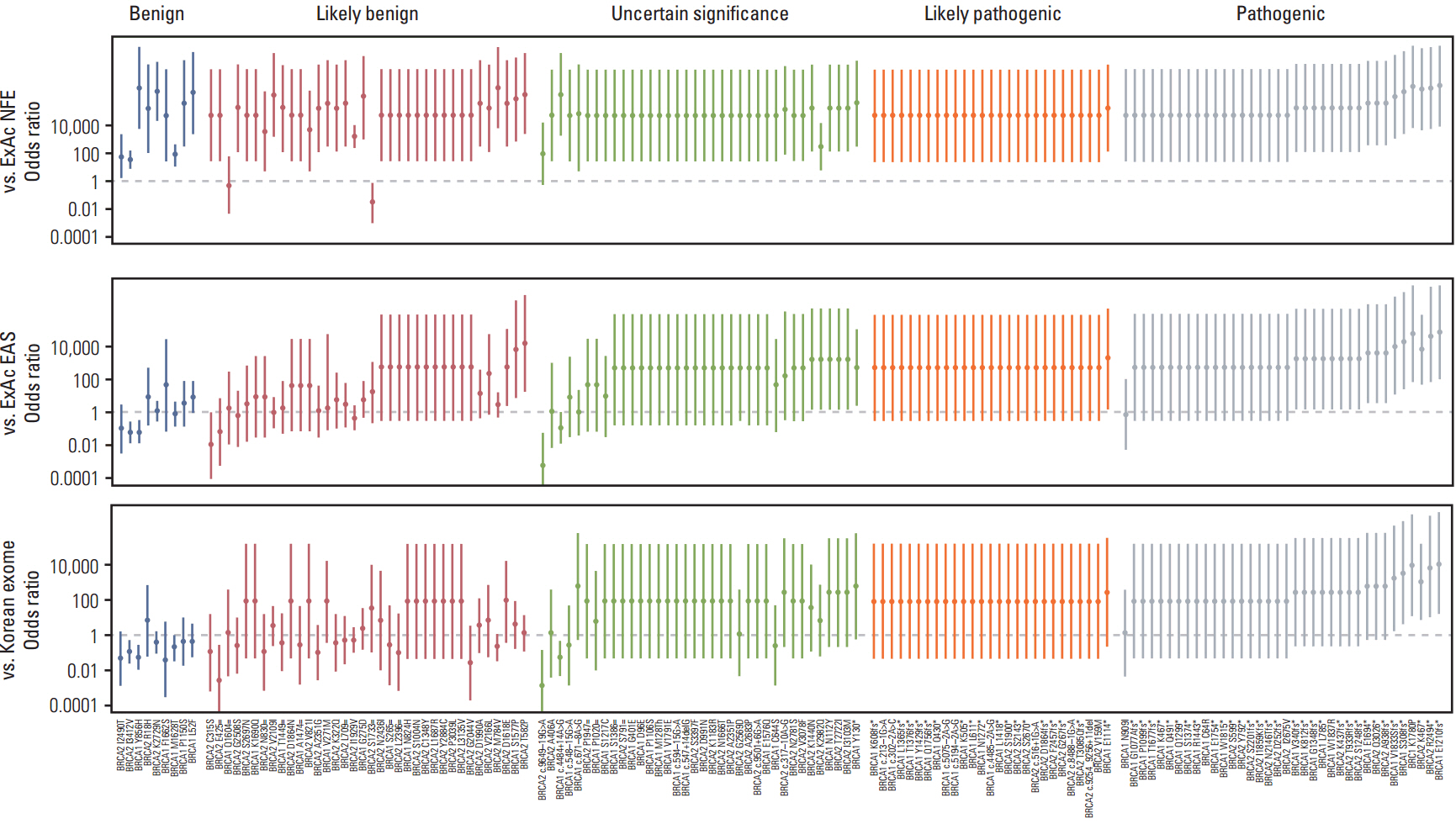
Comparison of variant frequencies in the general population has become an essential part of the American College of Medical Genetics and Genomics (ACMG) standards and guidelines for interpreting sequence variants. We determined the optimal number of relevant ethnic controls that should be used to accurately calculate the odds ratio (OR) of genetic variants.
MATERIALS AND METHODS
Using the ACMG guidelines, we reclassified BRCA1 and BRCA2 mutations and variants of unknown significance in 745 Korean patients susceptible to hereditary breast and ovarian cancer compared with 1,314 Korean population controls.
RESULTS
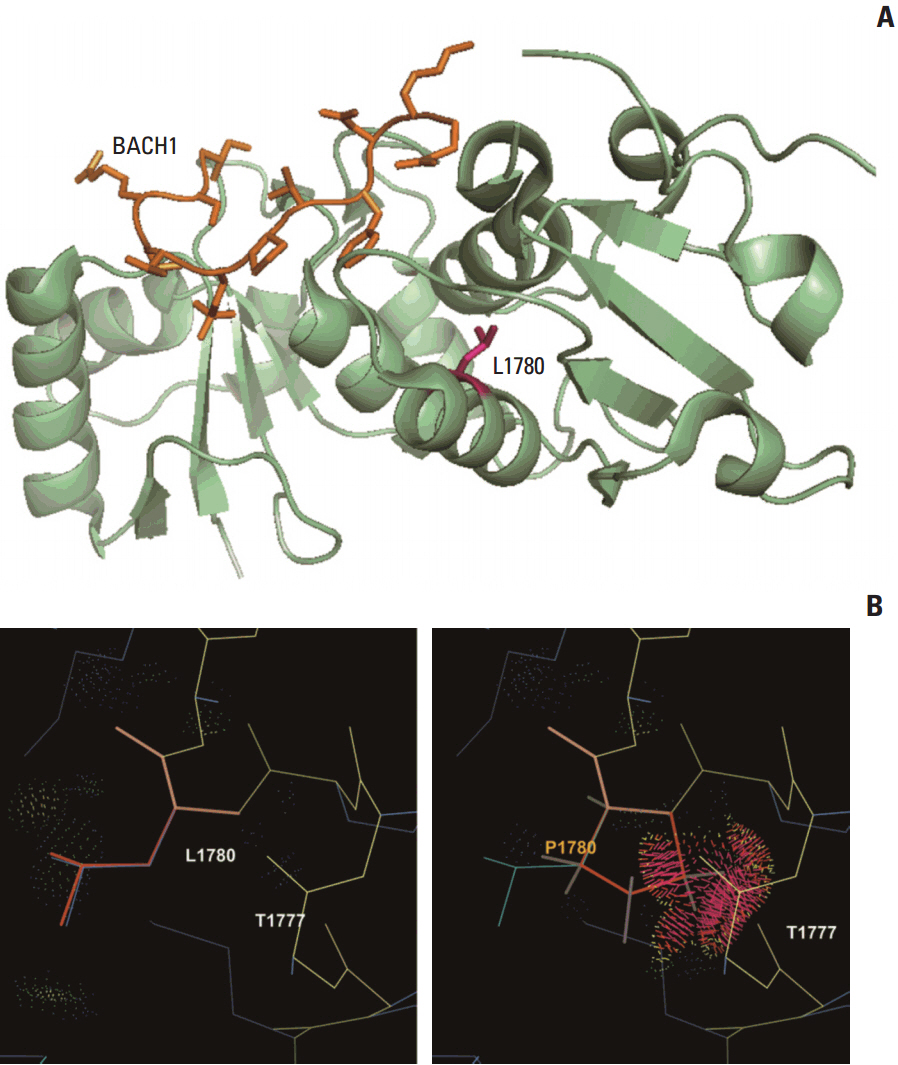
We observed that the ORs were falsely inflated when we analyzed several variants using non-Korean population data. Our simulation indicated that the number of controls needed for the lower limit of a 95% confidence interval to exceed 1.0 varied according to the frequency of the variant in each patient group, with more than 820 controls needed for a variant existing in 1% of cases. Using a sufficient number of relevant population data, we could efficiently classify variants and identified the BRCA1 p.Leu1780Pro mutation as a possible pathogenic founder mutation in Korean patients.
CONCLUSION
Our study suggests that BRCA1 p.Leu1780Pro is a novel pathogenic mutation found in Korean patients. We also determined the optimal number of relevant ethnic controls needed for accurate variant classification according to the ACMG guidelines.
Keyword
MeSH Terms
Figure
Cited by 2 articles
-
Clinical significance of variants of unknown significances in BRCA genes
Min Chul Choi
J Gynecol Oncol. 2019;30(4):. doi: 10.3802/jgo.2019.30.e80.Clinicopathological Features of Patients with the
BRCA1 c.5339T>C (p.Leu1780Pro) Variant
Hyung Seok Park, Jai Min Ryu, Ji Soo Park, Seock-Ah Im, So-Youn Jung, Eun-Kyu Kim, Woo-Chan Park, Jun Won Min, Jeeyeon Lee, Ji Young You, Jeong Eon Lee, Sung-Won Kim
Cancer Res Treat. 2020;52(3):680-688. doi: 10.4143/crt.2019.351.
Reference
-
References
1. Shiovitz S, Korde LA. Genetics of breast cancer: a topic in evolution. Ann Oncol. 2015; 26:1291–9.
Article2. Mersch J, Jackson MA, Park M, Nebgen D, Peterson SK, Singletary C, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. 2015; 121:269–75.3. Domchek SM, Bradbury A, Garber JE, Offit K, Robson ME. Multiplex genetic testing for cancer susceptibility: out on the high wire without a net? J Clin Oncol. 2013; 31:1267–70.
Article4. Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007; 81:873–83.
Article5. Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002; 20:1480–90.
Article6. Lindor NM, Goldgar DE, Tavtigian SV, Plon SE, Couch FJ. BRCA1/2 sequence variants of uncertain significance: a primer for providers to assist in discussions and in medical management. Oncologist. 2013; 18:518–24.
Article7. Ready K, Gutierrez-Barrera AM, Amos C, Meric-Bernstam F, Lu K, Hortobagyi G, et al. Cancer risk management decisions of women with BRCA1 or BRCA2 variants of uncertain significance. Breast J. 2011; 17:210–2.
Article8. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.
Article9. Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet. 2016; 98:1067–76.
Article10. Jang MA, Lee SH, Kim N, Ki CS. Frequency and spectrum of actionable pathogenic secondary findings in 196 Korean exomes. Genet Med. 2015; 17:1007–11.
Article11. Maxwell KN, Hart SN, Vijai J, Schrader KA, Slavin TP, Thomas T, et al. Evaluation of ACMG-guideline-based variant classification of cancer susceptibility and non-cancer-associated genes in families affected by breast cancer. Am J Hum Genet. 2016; 98:801–17.
Article12. Park KS, Cho EY, Nam SJ, Ki CS, Kim JW. Comparative analysis of BRCA1 and BRCA2 variants of uncertain significance in patients with breast cancer: a multifactorial probability-based model versus ACMG standards and guidelines for interpreting sequence variants. Genet Med. 2016; 18:1250–7.
Article13. Pagano M, Gauvreau K. Principles of biostatistics. 2nd ed. Pacific Grove, CA: Duxbury;2000.14. Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016; 37:235–41.
Article15. Jarvik GP, Browning BL. Consideration of cosegregation in the pathogenicity classification of genomic variants. Am J Hum Genet. 2016; 98:1077–81.
Article16. Chen VB, Davis IW, Richardson DC. KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci. 2009; 18:2403–9.
Article17. Petrucelli N, Daly MB, Feldman GL. Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet Med. 2010; 12:245–59.
Article18. Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994; 266:120–2.19. Shattuck-Eidens D, McClure M, Simard J, Labrie F, Narod S, Couch F, et al. A collaborative survey of 80 mutations in the BRCA1 breast and ovarian cancer susceptibility gene. Implications for presymptomatic testing and screening. JAMA. 1995; 273:535–41.
Article20. Cheon JY, Mozersky J, Cook-Deegan R. Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med. 2014; 6:121.
Article21. Eggington JM, Bowles KR, Moyes K, Manley S, Esterling L, Sizemore S, et al. A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin Genet. 2014; 86:229–37.
Article22. Choi DH, Lee MH, Bale AE, Carter D, Haffty BG. Incidence of BRCA1 and BRCA2 mutations in young Korean breast cancer patients. J Clin Oncol. 2004; 22:1638–45.
Article23. Han SH, Lee KR, Lee DG, Kim BY, Lee KE, Chung WS. Mutation analysis of BRCA1 and BRCA2 from 793 Korean patients with sporadic breast cancer. Clin Genet. 2006; 70:496–501.
Article24. Jang JH, Lee JE, Kwon MJ, Ki CS, Kim JW, Nam SJ, et al. Spectra of BRCA1 and BRCA2 mutations in Korean patients with breast cancer: the importance of whole-gene sequencing. J Hum Genet. 2012; 57:212–5.
Article25. Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010; 11:138–48.
Article26. Lee MS, Green R, Marsillac SM, Coquelle N, Williams RS, Yeung T, et al. Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res. 2010; 70:4880–90.
Article27. Spurdle AB, Whiley PJ, Thompson B, Feng B, Healey S, Brown MA, et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J Med Genet. 2012; 49:525–32.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Reclassification of BRCA1 and BRCA2 variants found in ovarian epithelial, fallopian tube, and primary peritoneal cancers
- Prevalence of BRCA1 and BRCA2 Germline Mutations in Breast Cancer Women of Multiple Ethnic Region in Northwest China
- Frequency of BRCA1 and BRCA2 Germline Mutations Detected by Protein Truncation Test and Cumulative Risks of Breast and Ovarian Cancer among Mutation Carriers in Japanese Breast Cancer Families
- BRCA1/BRCA2 Pathogenic Variant Breast Cancer: Treatment and Prevention Strategies
- Clinically Significant Unclassified Variants in BRCA1 and BRCA2 genes among Korean Breast Cancer Patients