Cancer Res Treat.
2017 Apr;49(2):387-398. 10.4143/crt.2016.106.
Underexpression of HOXA11 Is Associated with Treatment Resistance and Poor Prognosis in Glioblastoma
- Affiliations
-
- 1Department of Neurosurgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea. gknife@snu.ac.kr
- 2Department of Neurosurgery, Seoul National University College of Medicine, Seoul, Korea. nsckpark@snu.ac.kr
- 3Department of Physiology, Ajou University School of Medicine, Suwon, Korea.
- 4Department of Neurosurgery, Ajou University School of Medicine, Suwon, Korea.
- 5Department of Neurosurgery, Korea University College of Medicine, Seoul, Korea.
- 6Department of Radiation Oncology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea.
- 7Department of Internal Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea.
- 8Department of Neurology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea.
- 9Department of Radiology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea.
- 10Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea.
- KMID: 2378110
- DOI: http://doi.org/10.4143/crt.2016.106
Abstract
- PURPOSE
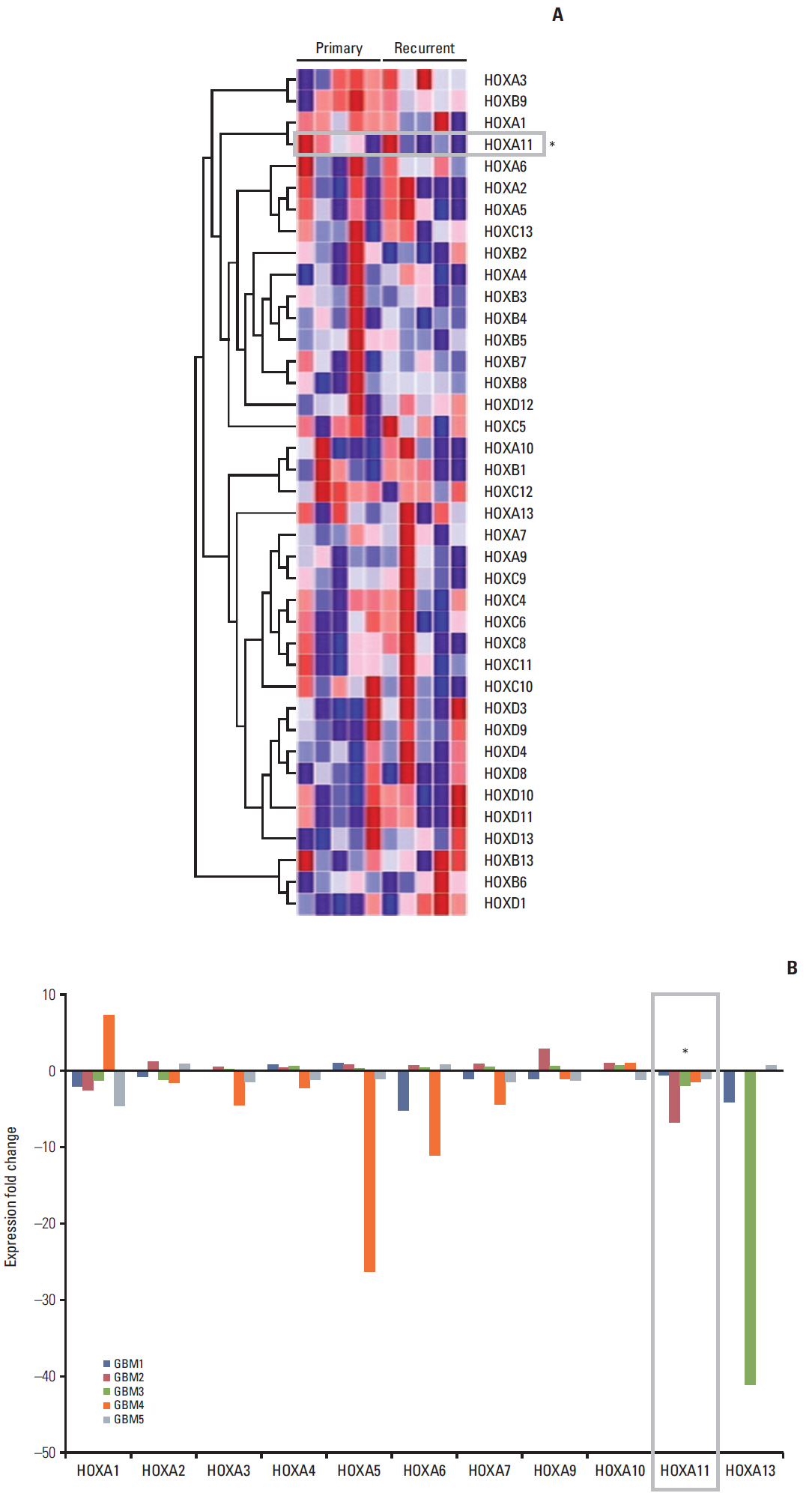
Homeobox (HOX) genes are essential developmental regulators that should normally be in the silenced state in an adult brain. The aberrant expression of HOX genes has been associated with the prognosis of many cancer types, including glioblastoma (GBM). This study examined the identity and role of HOX genes affecting GBM prognosis and treatment resistance.
MATERIALS AND METHODS
The full series of HOX genes of five pairs of initial and recurrent human GBM samples were screened by microarray analysis to determine the most plausible candidate responsible for GBM prognosis. Another 20 newly diagnosed GBM samples were used for prognostic validation. In vitro experiments were performed to confirm the role of HOX in treatment resistance. Mediators involved in HOX gene regulation were searched using differentially expressed gene analysis, gene set enrichment tests, and network analysis.
RESULTS
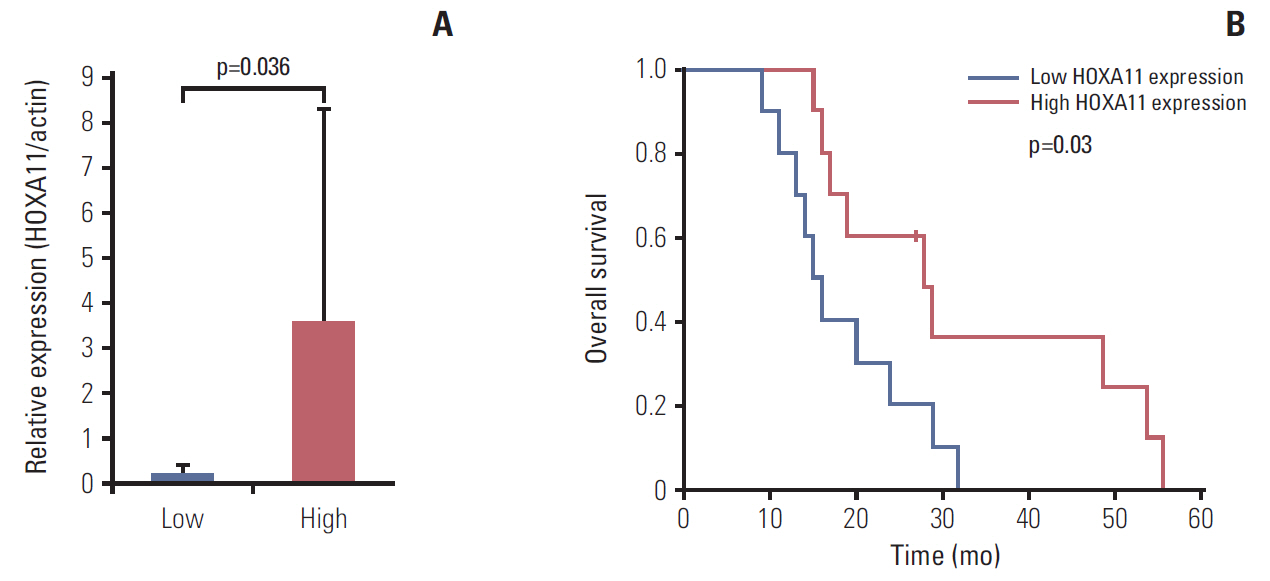
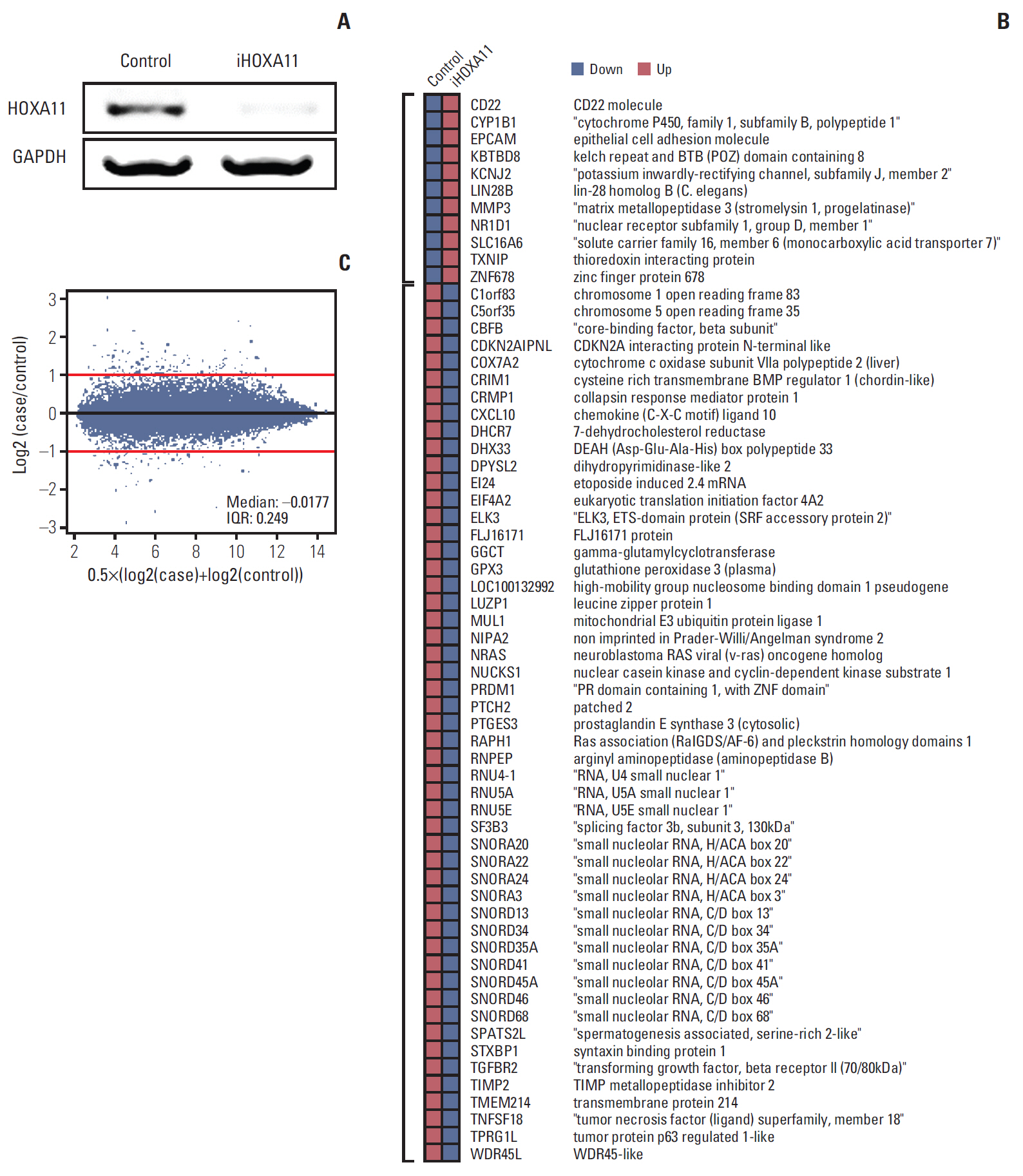
The underexpression of HOXA11 was identified as a consistent signature for a poor prognosis among the HOX genes. The overall survival of the GBM patients indicated a significantly favorable prognosis in patients with high HOXA11 expression (31±15.3 months) compared to the prognoses in thosewith low HOXA11 expression (18±7.3 months, p=0.03). When HOXA11 was suppressed in the GBM cell lines, the anticancer effect of radiotherapy and/or temozolomide declined. In addition, five candidate mediators (TGFBR2, CRIM1, TXNIP, DPYSL2, and CRMP1) that may confer an oncologic effect after HOXA11 suppression were identified.
CONCLUSION
The treatment resistance induced by the underexpression of HOXA11 can contribute to a poor prognosis in GBM. Further investigation will be needed to confirm the value of HOXA11 as a potential target for overcoming the treatment resistance by developing chemo- or radiosensitizers.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010; 10:361–71.
Article2. Takahashi Y, Hamada J, Murakawa K, Takada M, Tada M, Nogami I, et al. Expression profiles of 39 HOX genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative real-time RT-PCR system. Exp Cell Res. 2004; 293:144–53.
Article3. Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002; 2:777–85.
Article4. Abdel-Fattah R, Xiao A, Bomgardner D, Pease CS, Lopes MB, Hussaini IM. Differential expression of HOX genes in neoplastic and non-neoplastic human astrocytes. J Pathol. 2006; 209:15–24.5. Bodey B, Bodey B Jr, Siegel SE, Kaiser HE. Immunocytochemical detection of the homeobox B3, B4, and C6 gene products in childhood medulloblastomas/primitive neuroectodermal tumors. Anticancer Res. 2000; 20:1769–80.6. Buccoliero AM, Castiglione F, Rossi Degl'Innocenti D, Ammanati F, Giordano F, Sanzo M, et al. Hox-D genes expression in pediatric low-grade gliomas: real-time-PCR study. Cell Mol Neurobiol. 2009; 29:1–6.
Article7. Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003; 102:262–8.
Article8. Miao J, Wang Z, Provencher H, Muir B, Dahiya S, Carney E, et al. HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci U S A. 2007; 104:17093–8.
Article9. Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008; 26:3015–24.
Article10. Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, et al. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007; 13:6327–34.
Article11. Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010; 70:9243–52.12. Kim JW, Kim JY, Kim JE, Kim SK, Chung HT, Park CK. HOXA10 is associated with temozolomide resistance through regulation of the homologous recombinant DNA repair pathway in glioblastoma cell lines. Genes Cancer. 2014; 5:165–74.
Article13. Kim KJ, Moon SM, Kim SA, Kang KW, Yoon JH, Ahn SG. Transcriptional regulation of MDR-1 by HOXC6 in multidrug-resistant cells. Oncogene. 2013; 32:3339–49.
Article14. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006; 444:756–60.
Article15. Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012; 488:522–6.
Article16. Atkinson SP, Koch CM, Clelland GK, Willcox S, Fowler JC, Stewart R, et al. Epigenetic marking prepares the human HOXA cluster for activation during differentiation of pluripotent cells. Stem Cells. 2008; 26:1174–85.
Article17. Kwon SM, Kang SH, Park CK, Jung S, Park ES, Lee JS, et al. Recurrent glioblastomas reveal molecular subtypes associated with mechanistic implications of drug-resistance. PLoS One. 2015; 10:e0140528.
Article18. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006; 38:500–1.
Article19. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57.
Article20. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010; 38:W214–20.
Article21. Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008; 30:965–79.22. Ohgo S, Itoh A, Suzuki M, Satoh A, Yokoyama H, Tamura K. Analysis of hoxa11 and hoxa13 expression during patternless limb regeneration in Xenopus. Dev Biol. 2010; 338:148–57.
Article23. Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007; 26:6766–76.
Article24. Bahrani-Mostafavi Z, Tickle TL, Zhang J, Bennett KE, Vachris JC, Spencer MD, et al. Correlation analysis of HOX, ErbB and IGFBP family gene expression in ovarian cancer. Cancer Invest. 2008; 26:990–8.
Article25. Cillo C, Barba P, Freschi G, Bucciarelli G, Magli MC, Boncinelli E. HOX gene expression in normal and neoplastic human kidney. Int J Cancer. 1992; 51:892–7.
Article26. De Vita G, Barba P, Odartchenko N, Givel JC, Freschi G, Bucciarelli G, et al. Expression of homeobox-containing genes in primary and metastatic colorectal cancer. Eur J Cancer. 1993; 29A:887–93.
Article27. Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010; 70:453–62.28. Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, et al. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013; 19:7078–88.
Article29. Carrio M, Arderiu G, Myers C, Boudreau NJ. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer Res. 2005; 65:7177–85.
Article30. Petrini M, Felicetti F, Bottero L, Errico MC, Morsilli O, Boe A, et al. HOXB1 restored expression promotes apoptosis and differentiation in the HL60 leukemic cell line. Cancer Cell Int. 2013; 13:101.
Article31. Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol. 2005; 3:240–52.
Article32. Carrera M, Bitu CC, de Oliveira CE, Cervigne NK, Graner E, Manninen A, et al. HOXA10 controls proliferation, migration and invasion in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2015; 8:3613–23.33. Cui XP, Qin CK, Zhang ZH, Su ZX, Liu X, Wang SK, et al. HOXA10 promotes cell invasion and MMP-3 expression via TGFbeta2-mediated activation of the p38 MAPK pathway in pancreatic cancer cells. Dig Dis Sci. 2014; 59:1442–51.34. Kurscheid S, Bady P, Sciuscio D, Samarzija I, Shay T, Vassallo I, et al. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015; 16:16.
Article35. Li B, Cao X, Weng C, Wu Y, Fang X, Zhang X, et al. HoxA10 induces proliferation in human prostate carcinoma PC-3 cell line. Cell Biochem Biophys. 2014; 70:1363–8.
Article36. Liborio-Kimura TN, Jung HM, Chan EK. miR-494 represses HOXA10 expression and inhibits cell proliferation in oral cancer. Oral Oncol. 2015; 51:151–7.37. Oue N, Sentani K, Sakamoto N, Yasui W. Clinicopathologic and molecular characteristics of gastric cancer showing gastric and intestinal mucin phenotype. Cancer Sci. 2015; 106:951–8.
Article38. Xiao ZD, Jiao CY, Huang HT, He LJ, Zhao JJ, Lu ZY, et al. miR-218 modulate hepatocellular carcinoma cell proliferation through PTEN/AKT/PI3K pathway and HoxA10. Int J Clin Exp Pathol. 2014; 7:4039–44.39. Zhang HY, Li JH, Li G, Wang SR. Activation of ARK5/miR-1181/HOXA10 axis promotes epithelial-mesenchymal transition in ovarian cancer. Oncol Rep. 2015; 34:1193–202.
Article40. Zhang L, Wan Y, Jiang Y, Ma J, Liu J, Tang W, et al. Upregulation HOXA10 homeobox gene in endometrial cancer: role in cell cycle regulation. Med Oncol. 2014; 31:52.
Article41. Bai Y, Fang N, Gu T, Kang Y, Wu J, Yang D, et al. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int. 2014; 14:79.
Article42. Cui Y, Gao D, Linghu E, Zhan Q, Chen R, Brock MV, et al. Epigenetic changes and functional study of HOXA11 in human gastric cancer. Epigenomics. 2015; 7:201–13.
Article43. Fiegl H, Windbichler G, Mueller-Holzner E, Goebel G, Lechner M, Jacobs IJ, et al. HOXA11 DNA methylation: a novel prognostic biomarker in ovarian cancer. Int J Cancer. 2008; 123:725–9.44. Hwang JA, Lee BB, Kim Y, Park SE, Heo K, Hong SH, et al. HOXA11 hypermethylation is associated with progression of non-small cell lung cancer. Oncotarget. 2013; 4:2317–25.
Article45. Laffaire J, Everhard S, Idbaih A, Criniere E, Marie Y, de Reynies A, et al. Methylation profiling identifies 2 groups of gliomas according to their tumorigenesis. Neuro Oncol. 2011; 13:84–98.
Article46. Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, et al. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009; 4:255–64.
Article47. Skiriute D, Vaitkiene P, Asmoniene V, Steponaitis G, Deltuva VP, Tamasauskas A. Promoter methylation of AREG, HOXA11, hMLH1, NDRG2, NPTX2 and Tes genes in glioblastoma. J Neurooncol. 2013; 113:441–9.
Article48. Fang F, Munck J, Tang J, Taverna P, Wang Y, Miller DF, et al. The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer. Clin Cancer Res. 2014; 20:6504–16.
Article49. Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012; 72:2197–205.
Article50. Shih JY, Lee YC, Yang SC, Hong TM, Huang CY, Yang PC. Collapsin response mediator protein-1: a novel invasion-suppressor gene. Clin Exp Metastasis. 2003; 20:69–76.51. Mukherjee J, DeSouza LV, Micallef J, Karim Z, Croul S, Siu KW, et al. Loss of collapsin response mediator protein1, as detected by iTRAQ analysis, promotes invasion of human gliomas expressing mutant EGFRvIII. Cancer Res. 2009; 69:8545–54.
Article52. Yilmaz N, Ozaksit G, Terzi YK, Yilmaz S, Budak B, Aksakal O, et al. HOXA11 and MMP2 gene expression in uterosacral ligaments of women with pelvic organ prolapse. J Turk Ger Gynecol Assoc. 2014; 15:104–8.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Congenital Cerebral Glioblastoma Multiforme
- A Case of Intramedullary Glioblastoma Multiforme Involving Thoracic Cord in Child
- A Case of Glioblastoma Multiforme Arising in the Right Temporal Lobe in a Child
- Intramedullary Glioblastoma Multiforme: Report of 3 Cases
- Glioblastoma specific antigens, GD2 and CD90, are not involved in cancer stemness