Ann Lab Med.
2016 Nov;36(6):573-582. 10.3343/alm.2016.36.6.573.
Abnormalities in Chromosomes 1q and 13 Independently Correlate With Factors of Poor Prognosis in Multiple Myeloma
- Affiliations
-
- 1Department of Laboratory Medicine, Hallym University College of Medicine, Anyang, Korea. lyoungk@hallym.or.kr
- 2Department of Occupational and Environmental Medicine, Hallym University College of Medicine, Anyang, Korea.
- 3Department of Internal Medicine, Hallym University College of Medicine, Anyang, Korea.
- 4Department of Laboratory Medicine, Seoul National University College of Medicine, Seoul, Korea.
- 5Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea.
- KMID: 2373596
- DOI: http://doi.org/10.3343/alm.2016.36.6.573
Abstract
- BACKGROUND
We comprehensively profiled cytogenetic abnormalities in multiple myeloma (MM) and analyzed the relationship between cytogenetic abnormalities of undetermined prognostic significance and established prognostic factors.
METHODS
The karyotype of 333 newly diagnosed MM cases was analyzed in association with established prognostic factors. Survival analysis was also performed.
RESULTS
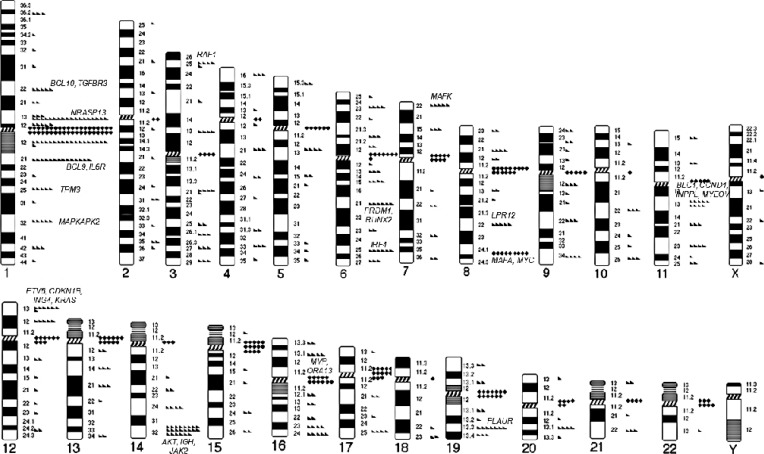
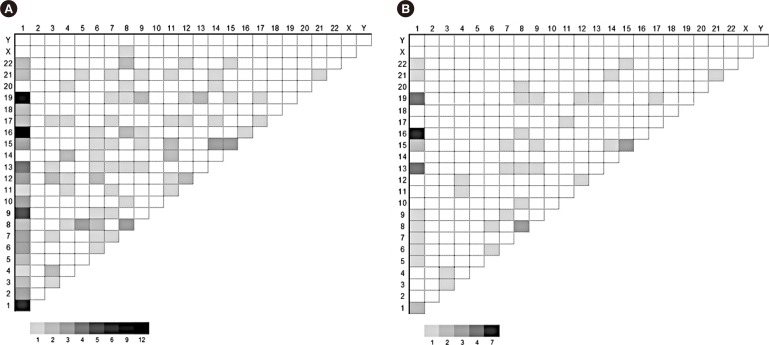
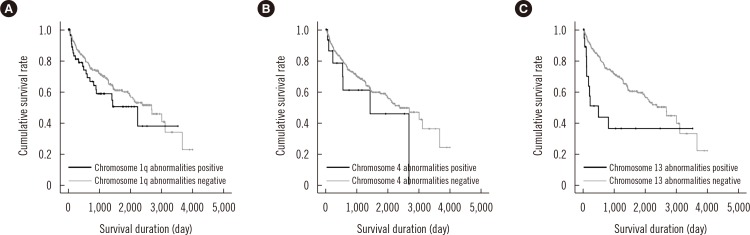
MM with abnormal karyotypes (41.1%) exhibited high international scoring system (ISS) stage, frequent IgA type, elevated IgG or IgA levels, elevated calcium levels, elevated creatine (Cr) levels, elevated β2-microglobulin levels, and decreased Hb levels. Structural abnormalities in chromosomes 1q, 4, and 13 were independently associated with elevated levels of IgG or IgA, calcium, and Cr, respectively. Chromosome 13 abnormalities were associated with poor prognosis and decreased overall survival.
CONCLUSIONS
This is the first study to demonstrate that abnormalities in chromosomes 1q, 4, and 13 are associated with established factors for poor prognosis, irrespective of the presence of other concurrent chromosomal abnormalities. Chromosome 13 abnormalities have a prognostic impact on overall survival in association with elevated Cr levels. Frequent centromeric breakpoints appear to be related to MM pathogenesis.
Keyword
MeSH Terms
-
Adolescent
Adult
Aged
Aged, 80 and over
Calcium/blood
*Chromosome Aberrations
Chromosomes, Human, Pair 1
Chromosomes, Human, Pair 13
Chromosomes, Human, Pair 4
Creatine/blood
Female
Hemoglobins/analysis
Humans
Immunoglobulin A/blood
Immunoglobulin G/blood
Karyotyping
Male
Middle Aged
Multiple Myeloma/*diagnosis/genetics/mortality
Multivariate Analysis
Prognosis
Survival Rate
Young Adult
Calcium
Creatine
Hemoglobins
Immunoglobulin A
Immunoglobulin G
Figure
Reference
-
1. Shaughnessy JD Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007; 109:2276–2284. PMID: 17105813.2. Segges P, Braggio E. Genetic markers used for risk stratification in multiple myeloma. Genet Res Int. 2011; 2011:798089. PMID: 22567368.3. Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005; 23:6333–6338. PMID: 16155016.4. Rajkumar SV. Multiple myeloma: 2013 update on diagnosis, risk-stratification, and management. Am J Hematol. 2013; 88:226–235. PMID: 23440663.5. Swerdlow SH, Campo E, et al. WHO classification of tumours of hematopoietic and lymphoid tissues. Lion: IARC Press;2008.6. Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995; 82:41–49. PMID: 7627933.7. Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006; 108:1724–1732. PMID: 16705089.8. Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009; 23:2210–2221. PMID: 19798094.9. Chng WJ, Santana-Dávila R, Van Wier SA, Ahmann GJ, Jalal SM, Bergsagel PL, et al. Prognostic factors for hyperdiploid-myeloma: effects of chromosome 13 deletions and IgH translocations. Leukemia. 2006; 20:807–813. PMID: 16511510.10. Avet-Louseau H, Daviet A, Sauner S, Bataille R. Intergroupe Francophone du Myélome. Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br J Haematol. 2000; 111:1116–1117. PMID: 11227093.11. Lodé L, Eveillard M, Trichet V, Soussi T, Wuillème S, Richebourg S, et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica. 2010; 95:1973–1976. PMID: 20634494.12. Munshi NC, Anderson KC, Bergsagel PL, Shaughnessy J, Palumbo A, Durie B, et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011; 117:4696–4700. PMID: 21292777.13. Marzin Y, Jamet D, Douet-Guilbert N, Morel F, Le Bris MJ, Morice P, et al. Chromosome 1 abnormalities in multiple myeloma. Anticancer Res. 2006; 26:953–959. PMID: 16619492.14. Wu KL, Beverloo B, Lokhorst HM, Segeren CM, van der Holt B, Steijaert MM, et al. Abnormalities of chromosome 1p/q are highly associated with chromosome 13/13q deletions and are an adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Br J Haematol. 2007; 136:615–623. PMID: 17223915.15. Boyd KD, Ross FM, Walker BA, Wardell CP, Tapper WJ, Chiecchio L, et al. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res. 2011; 17:7776–7784. PMID: 21994415.16. Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010; 116:e56–e65. PMID: 20616218.17. Zhan F, Colla S, Wu X, Chen B, Stewart JP, Kuehl WM, et al. CKS1B, overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and-independent mechanisms. Blood. 2007; 109:4995–5001. PMID: 17303695.18. Poretti G, Kwee I, Bernasconi B, Rancoita PM, Rinaldi A, Capella C, et al. Chromosome 11q23.1 is an unstable region in B-cell tumor cell lines. Leuk Res. 2011; 35:808–813. PMID: 21420167.19. Hallek M, Bergsagel PL, Anderson KC. Multiple myeloma: increasing evidence for a multistep transformation process. Blood. 1998; 91:3–21. PMID: 9414264.20. Shaffer LG, Tommerup N, editors. ISCN 2005: an international system for human cytogenetic nomenclature. Basel, Switzerland: Karger Medical and Scientific Publishers;2005.21. Shaffer LG, Slovak ML, editors. ISCN 2009: an international system for human cytogenetic nomenclature. Basel, Switzerland: Karger Medical and Scientific Publishers;2009.22. Shaffer LG, McGowan-Jordan J, editors. ISCN 2013: an international system for human cytogenetic nomenclature. Basel, Switzerland: Karger Medical and Scientific Publishers;2013.23. Durie BG, Salmon SE. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer. 1975; 36:842–854. PMID: 1182674.24. Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Bladé J, et al. International staging system for multiple myeloma. J Clin Oncol. 2005; 23:3412–3420. PMID: 15809451.25. Drayson M, Begum G, Basu S, Makkuni S, Dunn J, Barth N, et al. Effects of paraprotein heavy and light chain types and free light chain load on survival in myeloma: an analysis of patients receiving conventional-dose chemotherapy in Medical Research Council UK multiple myeloma trials. Blood. 2006; 108:2013–2019. PMID: 16728700.26. Bradwell A, Harding S, Fourrier N, Mathiot C, Attal M, Moreau P, et al. Prognostic utility of intact immunoglobulin Ig'κ/Ig'λ ratios in multiple myeloma patients. Leukemia. 2013; 27:202–207. PMID: 22699454.27. Oh S, Koo DH, Kwon MJ, Kim K, Suh C, Min CK, et al. Chromosome 13 deletion and hypodiploidy on conventional cytogenetics are robust prognostic factors in Korean multiple myeloma patients: web-based multicenter registry study. Ann Hematol. 2014; 93:1353–1361. PMID: 24671365.28. Cigudosa JC, Rao PH, Calasanz MJ, Odero MD, Michaeli J, Jhanwar SC, et al. Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood. 1998; 91:3007–3010. PMID: 9531613.29. Kim S, Kim H, Kang H, Kim J, Eom H, Kim T, et al. Clinical significance of cytogenetic aberrations in bone marrow of patients with diffuse large B-cell lymphoma: prognostic significance and relevance to histologic involvement. J Hematol Oncol. 2013; 6:76. PMID: 24220305.30. Sawyer JR, Tricot G, Mattox S, Jagannath S, Barlogie B. Jumping translocations of chromosome 1q in multiple myeloma: evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood. 1998; 91:1732–1741. PMID: 9473240.31. Lauta VM. Interleukin-6 and the network of several cytokines in multiple myeloma: an overview of clinical and experimental data. Cytokine. 2001; 16:79–86. PMID: 11741345.32. Lamant L, Dastugue N, Pulford K, Delsol G, Mariamé B. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood. 1999; 93:3088–3095. PMID: 10216106.33. Felix RS, Colleoni GW, Caballero OL, Yamamoto M, Almeida MS, Andrade VC, et al. SAGE analysis highlights the importance of p53csv, ddx5, mapkapk2 and ranbp2 to multiple myeloma tumorigenesis. Cancer Lett. 2009; 278:41–48. PMID: 19171422.34. Nahi H, Sutlu T, Jansson M, Alici E, Gahrton G. Clinical impact of chromosomal aberrations in multiple myeloma. J Intern Med. 2011; 269:137–147. PMID: 21158983.35. Smadja NV, Fruchart C, Isnard F, Louvet C, Dutel JL, Cheron N, et al. Chromosomal analysis in multiple myeloma: cytogenetic evidence of two different diseases. Leukemia. 1998; 12:960–969. PMID: 9639426.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correlation of Chromosomal Aberrations with Prognostic Markers in Multiple Myeloma Patients- A Single Institution Study
- Rapid Progression of Solitary Plasmacytoma to Multiple Myeloma in Lumbar Vertebra
- A Case of Cutaneous Plasmacytoma Treated with Thalidomide
- Clinical Utility of a Diagnostic Approach to Detect Genetic Abnormalities in Multiple Myeloma: A Single Institution Experience
- Salmonella Pyomyositisina Multiple Myeloma Patient: A Case Report