Ann Lab Med.
2016 May;36(3):255-258. 10.3343/alm.2016.36.3.255.
Genotyping Influenza Virus by Next-Generation Deep Sequencing in Clinical Specimens
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea. sparkle@snu.ac.kr
- KMID: 2373536
- DOI: http://doi.org/10.3343/alm.2016.36.3.255
Abstract
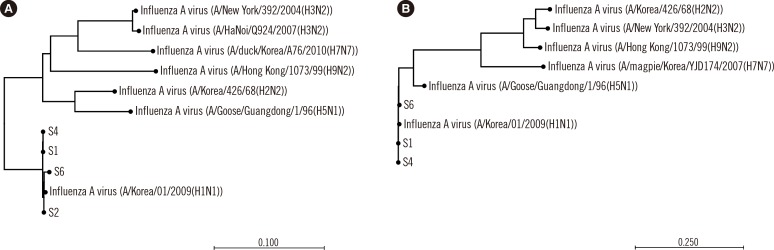
- Rapid and accurate identification of an influenza outbreak is essential for patient care and treatment. We describe a next-generation sequencing (NGS)-based, unbiased deep sequencing method in clinical specimens to investigate an influenza outbreak. Nasopharyngeal swabs from patients were collected for molecular epidemiological analysis. Total RNA was sequenced by using the NGS technology as paired-end 250 bp reads. Total of 7 to 12 million reads were obtained. After mapping to the human reference genome, we analyzed the 3-4% of reads that originated from a non-human source. A BLAST search of the contigs reconstructed de novo revealed high sequence similarity with that of the pandemic H1N1 virus. In the phylogenetic analysis, the HA gene of our samples clustered closely with that of A/Senegal/VR785/2010(H1N1), A/Wisconsin/11/2013(H1N1), and A/Korea/01/2009(H1N1), and the NA gene of our samples clustered closely with A/Wisconsin/11/2013(H1N1). This study suggests that NGS-based unbiased sequencing can be effectively applied to investigate molecular characteristics of nosocomial influenza outbreak by using clinical specimens such as nasopharyngeal swabs.
Keyword
MeSH Terms
-
Databases, Genetic
Genotype
High-Throughput Nucleotide Sequencing
Humans
Influenza A Virus, H1N1 Subtype/classification/*genetics/isolation & purification
Influenza, Human/diagnosis/*virology
Nasopharynx/*virology
Nucleic Acid Amplification Techniques
Phylogeny
RNA, Viral/analysis/metabolism
Sequence Analysis, RNA
Viral Proteins/genetics
RNA, Viral
Viral Proteins
Figure
Reference
-
1. Bearden A, Friedrich TC, Goldberg TL, Byrne B, Spiegel C, Schult P, et al. An outbreak of the 2009 influenza a (H1N1) virus in a children's hospital. Influenza Other Respir Viruses. 2012; 6:374–379. PMID: 22212690.
Article2. Pollara CP, Piccinelli G, Rossi G, Cattaneo C, Perandin F, Corbellini S, et al. Nosocomial outbreak of the pandemic Influenza A (H1N1) 2009 in critical hematologic patients during seasonal influenza 2010-2011: detection of oseltamivir resistant variant viruses. BMC Infect Dis. 2013; 13:127. PMID: 23496867.
Article3. Tsagris V, Nika A, Kyriakou D, Kapetanakis I, Harahousou E, Stripeli F, et al. Influenza A/H1N1/2009 outbreak in a neonatal intensive care unit. J Hosp Infect. 2012; 81:36–40. PMID: 22463979.
Article4. Mengelle C, Mansuy JM, Da Silva I, Guerin JL, Izopet J. Evaluation of a polymerase chain reaction-electrospray ionization time-of-flight mass spectrometry for the detection and subtyping of influenza viruses in respiratory specimens. J Clin Virol. 2013; 57:222–226. PMID: 23557709.
Article5. Munro SB, Kuypers J, Jerome KR. Comparison of a multiplex real-time PCR assay with a multiplex Luminex assay for influenza virus detection. J Clin Microbiol. 2013; 51:1124–1129. PMID: 23345299.
Article6. Ramakrishnan MA, Tu ZJ, Singh S, Chockalingam AK, Gramer MR, Wang P, et al. The feasibility of using high resolution genome sequencing of influenza A viruses to detect mixed infections and quasispecies. PLoS One. 2009; 4:e7105. PMID: 19771155.
Article7. Renzette N, Caffrey DR, Zeldovich KB, Liu P, Gallagher GR, Aiello D, et al. Evolution of the influenza A virus genome during development of oseltamivir resistance in vitro. J Virol. 2014; 88:272–281. PMID: 24155392.
Article8. Sherry NL, Porter JL, Seemann T, Watkins A, Stinear TP, Howden BP. Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J Clin Microbiol. 2013; 51:1396–1401. PMID: 23408689.
Article9. Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013; 13:137–146. PMID: 23158499.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prevention and Treatment of Influenza
- Benign Acute Childhood Myositis Associated with Influenza B Virus
- Progress and hurdles in the development of influenza virus-like particle vaccines for veterinary use
- The Comparison of Restriction Fragment Mass Polymorphism with Sequencing Method for the Hepatitis C Virus Genotyping
- Epidemiological Analysis of Influenza by Laboratory Surveillance in Incheon, 2003/2004~2004/2005