Healthc Inform Res.
2012 Dec;18(4):272-278. 10.4258/hir.2012.18.4.272.
Development of Korean Rare Disease Knowledge Base
- Affiliations
-
- 1Seoul National University Biomedical Informatics (SNUBI), Division of Biomedical Informatics, Seoul National University College of Medicine, Seoul, Korea. juhan@snu.ac.kr
- 2Systems Biomedical Informatics Research Center, Seoul National University, Seoul, Korea.
- 3Department of Pediatrics, Seoul National University College of Medicine, Seoul, Korea.
- 4Pediatric Clinical Neuroscience Center, Seoul National University Children's Hospital, Seoul, Korea.
- 5Department of Pediatrics, Seoul National University Children's Hospital, Seoul, Korea.
- 6Research Center for Rare Diseases, Seoul National University Hospital, Seoul, Korea.
- 7Kidney Research Institute, Medical Research Center, Seoul National University College of Medicine, Seoul, Korea.
- KMID: 2284562
- DOI: http://doi.org/10.4258/hir.2012.18.4.272
Abstract
OBJECTIVES
Rare disease research requires a broad range of disease-related information for the discovery of causes of genetic disorders that are maladies caused by abnormalities in genes or chromosomes. A rarity in cases makes it difficult for researchers to elucidate definite inception. This knowledge base will be a major resource not only for clinicians, but also for the general public, who are unable to find consistent information on rare diseases in a single location.
METHODS
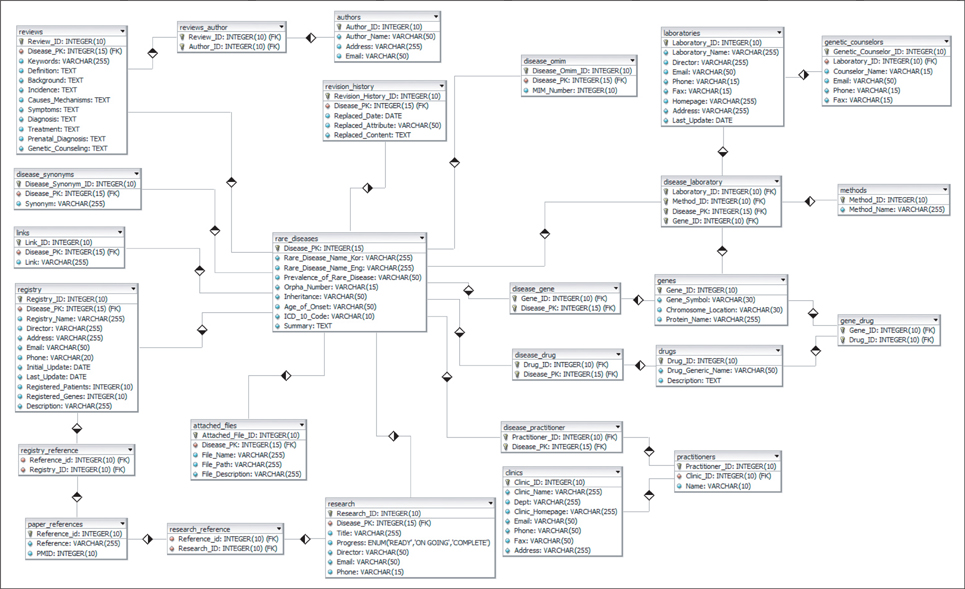
We design a compact database schema for faster querying; its structure is optimized to store heterogeneous data sources. Then, clinicians at Seoul National University Hospital (SNUH) review and revise those resources. Additionally, we integrated other sources to capture genomic resources and clinical trials in detail on the Korean Rare Disease Knowledge base (KRDK).
RESULTS
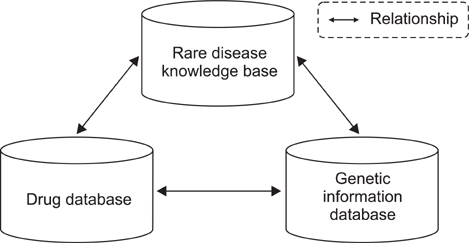
As a result, we have developed a Web-based knowledge base, KRDK, suitable for study of Mendelian diseases that commonly occur among Koreans. This knowledge base is comprised of disease summary and review, causal gene list, laboratory and clinic directory, patient registry, and so on. Furthermore, database for analyzing and giving access to human biological information and the clinical trial management system are integrated on KRDK.
CONCLUSIONS
We expect that KRDK, the first rare disease knowledge base in Korea, may contribute to collaborative research and be a reliable reference for application to clinical trials. Additionally, this knowledge base is ready for querying of drug information so that visitors can search a list of rare diseases that is relative to specific drugs. Visitors can have access to KRDK via http://www.snubi.org/software/raredisease/.
MeSH Terms
Figure
Reference
-
1. Hampton T. Rare disease research gets boost. JAMA. 2006. 295(24):2836–2838.
Article2. Pescucci C, Mari F, Longo I, Vogiatzi P, Caselli R, Scala E, et al. Autosomal-dominant Alport syndrome: natural history of a disease due to COL4A3 or COL4A4 gene. Kidney Int. 2004. 65(5):1598–1603.
Article3. Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009. 106(45):19096–19101.
Article4. Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010. 42(1):30–35.
Article5. EURORDIS. Rare diseases: understanding this public health priority [Internet]. 2005. cited at 2012 Dec 1. Paris: EURORDIS;Available from: http://www.eurordis.org/IMG/pdf/princeps_document-EN.pdf.6. Morgan SS, Aslam MB, Mukkanna KS, Ampat G. A rare presentation of sarcoidosis, back pain and spondylolisthesis. J Bone Joint Surg Br. 2008. 90(2):240–242.
Article7. Palo JU, Ulmanen I, Lukka M, Ellonen P, Sajantila A. Genetic markers and population history: Finland revisited. Eur J Hum Genet. 2009. 17(10):1336–1346.
Article8. Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat. 2002. 19(5):501–509.
Article9. Pagon RA. GeneReviews. 1993. Seattle (WA): University of Washington.10. Weinreich SS, Mangon R, Sikkens JJ, Teeuw ME, Cornel MC. Orphanet: a European database for rare diseases. Ned Tijdschr Geneeskd. 2008. 152(9):518–519.11. Park YR, Bae YJ, Kim JH. BioEMR: an integrative framework for cancer research with multiple genomic technologies. Summit on Translat Bioinforma. 2008. 2008:81–84.12. Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009. 461(7261):272–276.
Article13. Erlich Y, Edvardson S, Hodges E, Zenvirt S, Thekkat P, Shaag A, et al. Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res. 2011. 21(5):658–664.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Development of a Nursing Diagnostic Decision Support System for Inpatients with Type 2 Diabetes Mellitus Using Knowledge Engineering
- Development of a Knowledge Base for Korean Pharmacogenomics Research Network
- Design and Construction of a NLP Based Knowledge Extraction Methodology in the Medical Domain Applied to Clinical Information
- Establishment of Transsphenoidal Approach in Skull Base Surgery: A Historical Analysis
- A Case of Massive Skull Base Erosion by Fungus Ball in the Sphenoid Sinus