Allergy Asthma Immunol Res.
2014 Nov;6(6):496-503. 10.4168/aair.2014.6.6.496.
Novel genes in Human Asthma Based on a Mouse Model of Allergic Airway Inflammation and Human Investigations
- Affiliations
-
- 1Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary. szalaics@gmail.com
- 2Department of Pulmonology, Semmelweis University, Budapest, Hungary.
- 3Department of Cardiovascular Science, University of Sheffield, Sheffield, UK.
- 4Heim, Pal Children Hospital, Budapest, Hungary.
- 5Ministry of National Resources, Budapest, Hungary.
- 6Department of Measurement and Information Systems, University of Technology and Economics, Budapest, Hungary.
- 7Csertex Research Laboratory, Budapest, Hungary.
- KMID: 2260171
- DOI: http://doi.org/10.4168/aair.2014.6.6.496
Abstract
- PURPOSE
Based on a previous gene expression study in a mouse model of asthma, we selected 60 candidate genes and investigated their possible roles in human asthma.
METHODS
In these candidate genes, 90 SNPs were genotyped using MassARRAY technology from 311 asthmatic children and 360 healthy controls of the Hungarian (Caucasian) population. Moreover, gene expression levels were measured by RT PCR in the induced sputum of 13 asthmatics and 10 control individuals. t-tests, chi-square tests, and logistic regression were carried out in order to assess associations of SNP frequency and expression level with asthma. Permutation tests were performed to account for multiple hypothesis testing.
RESULTS
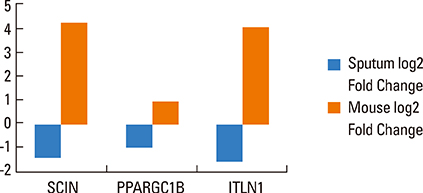
The frequency of 4 SNPs in 2 genes differed significantly between asthmatic and control subjects: SNPs rs2240572, rs2240571, rs3735222 in gene SCIN, and rs32588 in gene PPARGC1B. Carriers of the minor alleles had reduced risk of asthma with an odds ratio of 0.64 (0.51-0.80; P=7x10(-5)) in SCIN and 0.56 (0.42-0.76; P=1.2x10(-4)) in PPARGC1B. The expression levels of SCIN, PPARGC1B and ITLN1 genes were significantly lower in the sputum of asthmatics.
CONCLUSIONS
Three potentially novel asthma-associated genes were identified based on mouse experiments and human studies.
MeSH Terms
Figure
Reference
-
1. Holgate ST. Pathogenesis of asthma. Clin Exp Allergy. 2008; 38:872–897.2. Lemanske RF Jr, Busse WW. Asthma: clinical expression and molecular mechanisms. J Allergy Clin Immunol. 2010; 125:S95–S102.3. Braman SS. The global burden of asthma. Chest. 2006; 130:4S–12S.4. Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008; 8:169–182.5. Bierbaum S, Heinzmann A. The genetics of bronchial asthma in children. Respir Med. 2007; 101:1369–1375.6. Finkelman FD, Vercelli D. Advances in asthma, allergy mechanisms, and genetics in 2006. J Allergy Clin Immunol. 2007; 120:544–550.7. Weiss ST, Raby BA, Rogers A. Asthma genetics and genomics 2009. Curr Opin Genet Dev. 2009; 19:279–282.8. Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, Hamid Q, Rothenberg ME. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003; 111:1863–1874.9. Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Köhl J, Wahl L, Kuperman D, Germer S, Aud D, Peltz G, Wills-Karp M. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol. 2000; 1:221–226.10. Tölgyesi G, Molnár V, Semsei AF, Kiszel P, Ungvári I, Pócza P, Wiener Z, Komlósi ZI, Kunos L, Gálffy G, Losonczy G, Seres I, Falus A, Szalai C. Gene expression profiling of experimental asthma reveals a possible role of paraoxonase-1 in the disease. Int Immunol. 2009; 21:967–975.11. Di Valentin E, Crahay C, Garbacki N, Hennuy B, Guéders M, Noël A, Foidart JM, Grooten J, Colige A, Piette J, Cataldo D. New asthma biomarkers: lessons from murine models of acute and chronic asthma. Am J Physiol Lung Cell Mol Physiol. 2009; 296:L185–L197.12. Ungvári I, Hadadi E, Virág V, Bikov A, Nagy A, Semsei AF, Gálffy G, Tamási L, Horváth I, Szalai C. Implication of BIRC5 in asthma pathogenesis. Int Immunol. 2012; 24:293–301.13. Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haus-sler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496.14. International HapMap Project [Internet]. [place unknown]: International HapMap Project;cited 2013 May 31. Available from: http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36/.15. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21:263–265.16. Ungvári I, Hullám G, Antal P, Kiszel PS, Gézsi A, Hadadi É, Virág V, Hajós G, Millinghoffer A, Nagy A, Kiss A, Semsei ÁF, Temesi G, Melegh B, Kisfali P, Széll M, Bikov A, Gálffy G, Tamási L, Falus A, Szalai C. Evaluation of a partial genome screening of two asthma susceptibility regions using bayesian network based bayesian multilevel analysis of relevance. PLoS One. 2012; 7:e33573.17. Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, Adams OD, Macedo P, Booton R, Gibeon D, Chung KF, Lindsay MA. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012; 129:95–103.18. Tsaparas P, Mariño-Ramírez L, Bodenreider O, Koonin EV, Jordan IK. Global similarity and local divergence in human and mouse gene co-expression networks. BMC Evol Biol. 2006; 6:70.19. Kuhn A, Luthi-Carter R, Delorenzi M. Cross-species and cross-platform gene expression studies with the Bioconductor-compliant R package 'annotationTools'. BMC Bioinformatics. 2008; 9:26.20. Strand AD, Aragaki AK, Baquet ZC, Hodges A, Cunningham P, Holmans P, Jones KR, Jones L, Kooperberg C, Olson JM. Conservation of regional gene expression in mouse and human brain. PLoS Genet. 2007; 3:e59.21. Song KH, Chiang JY. Glucagon and cAMP inhibit cholesterol 7alpha-hydroxylase (CYP7A1) gene expression in human hepatocytes: discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology. 2006; 43:117–125.22. Jordan IK, Mariño-Ramírez L, Koonin EV. Evolutionary significance of gene expression divergence. Gene. 2005; 345:119–126.23. Jordan IK, Mariño-Ramírez L, Wolf YI, Koonin EV. Conservation and coevolution in the scale-free human gene coexpression network. Mol Biol Evol. 2004; 21:2058–2070.24. Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002; 99:4465–4470.25. Yanai I, Graur D, Ophir R. Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. OMICS. 2004; 8:15–24.26. Ehre C, Rossi AH, Abdullah LH, De Pestel K, Hill S, Olsen JC, Davis CW. Barrier role of actin filaments in regulated mucin secretion from airway goblet cells. Am J Physiol Cell Physiol. 2005; 288:C46–C56.27. Davis CW, Dickey BF. Regulated airway goblet cell mucin secretion. Annu Rev Physiol. 2008; 70:487–512.28. Bush WS, McCauley JL, DeJager PL, Dudek SM, Hafler DA, Gibson RA, Matthews PM, Kappos L, Naegelin Y, Polman CH, Hauser SL, Oksenberg J, Haines JL, Ritchie MD. International Multiple Sclerosis Genetics Consortium. A knowledge-driven interaction analysis reveals potential neurodegenerative mechanism of multiple sclerosis susceptibility. Genes Immun. 2011; 12:335–340.29. Park SJ, Lee YC. Peroxisome proliferator-activated receptor gamma as a novel therapeutic target in asthma. J Asthma. 2008; 45:1–8.30. Lee KS, Park SJ, Hwang PH, Yi HK, Song CH, Chai OH, Kim JS, Lee MK, Lee YC. PPAR-gamma modulates allergic inflammation thro-ugh up-regulation of PTEN. FASEB J. 2005; 19:1033–1035.31. Lee SH, Jang AS, Woo Park S, Park JS, Kim YK, Uh ST, Kim YH, Chung IY, Park BL, Shin HD, Park CS. Genetic effect of single-nucleotide polymorphisms in the PPARGC1B gene on airway hyperreactivity in asthmatic patients. Clin Exp Allergy. 2011; 41:1533–1544.32. Tölgyesi G, Keszei M, Ungvári I, Nagy A, Falus A, Szalai C. Involvement of TNFalpha -308A promoter polymorphism in the development of asthma in children infected with Chlamydophila pneumoniae. Pediatr Res. 2006; 60:543–548.33. Ungvári I, Tölgyesi G, Semsei AF, Nagy A, Radosits K, Keszei M, Kozma GT, Falus A, Szalai C. CCR5 Delta 32 mutation, Mycoplasma pneumoniae infection, and asthma. J Allergy Clin Immunol. 2007; 119:1545–1547.34. Nagy A, Kozma GT, Keszei M, Treszl A, Falus A, Szalai C. The development of asthma in children infected with Chlamydia pneumoniae is dependent on the modifying effect of mannose-binding lectin. J Allergy Clin Immunol. 2003; 112:729–734.35. Nagy A, Keszei M, Kis Z, Budai I, Tölgyesi G, Ungvári I, Falus A, Szalai C. Chlamydophila pneumoniae infection status is dependent on the subtypes of asthma and allergy. Allergy Asthma Proc. 2007; 28:58–63.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Understanding the Mouse Model of Respiratory Allergic Diseases
- Evaluation of Human MSCs Treatment Frequency on Airway Inflammation in a Mouse Model of Acute Asthma
- Understanding asthma using animal models
- Ampicillin treated German cockroach extract leads to reduced inflammation in human lung cells and a mouse model of Asthma
- An Intratracheal Challenge Murine Model of Asthma: Can Bronchial Inflammation Affect the Nose?