Allergy Asthma Immunol Res.
2012 May;4(3):132-136. 10.4168/aair.2012.4.3.132.
Genetic Variations in TXNRD1 as Potential Predictors of Drug-Induced Liver Injury
- Affiliations
-
- 1Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Korea. guinea71@snu.ac.kr
- 2Institute of Allergy and Clinical Immunology, Seoul National University Medical Research Center, Seoul, Korea.
- 3DNA Link inc., Seoul, Korea.
- 4Department of Internal Medicine, Hanyang University College of Medicine, Seoul, Korea.
- 5Department of Internal Medicine, Eulji University College of Medicine, Seoul, Korea.
- 6Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Korea.
- 7Department of Life Science, Pohang University of Science and Technology, Pohang, Korea.
- 8Department of Allergy and Clinical Immunology, Ajou University School of Medicine, Suwon, Korea.
- KMID: 2167064
- DOI: http://doi.org/10.4168/aair.2012.4.3.132
Abstract
- PURPOSE
Drug-induced liver injury (DILI) is the most common adverse drug reaction; however, it is not easily predicted. We hypothesize that DILI has a common genetic basis. Based on the findings of previous animal studies on toxic hepatitis, we selected the thioredoxin reductase 1 gene (TXNRD1) as a candidate marker of DILI for this genetic association study.
METHODS
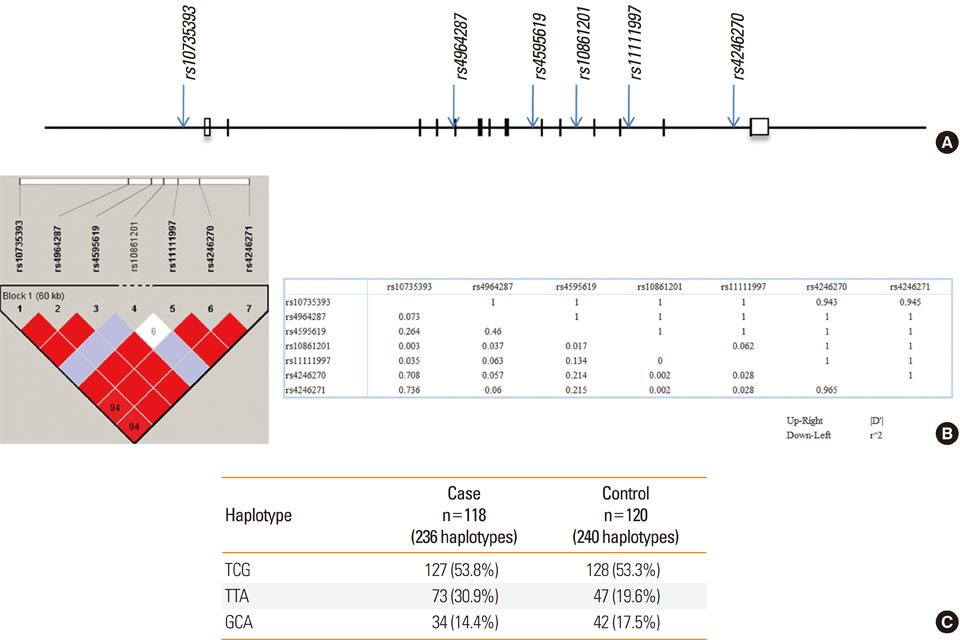
Records from 118 patients with DILI were extracted from the database of the Adverse Drug Reaction Research Group in South Korea. Causative drugs included antituberculosis drugs (n=68, 57.6%), antibiotics (n=22, 18.6%), antiepileptic drugs (n=7, 5.9%), non-steroidal anti-inflammatory drugs (n=5, 4.2%), and others (n=16, 13.7%). Seven single nucleotide polymorphisms (SNPs) in TXNRD1 (rs10735393, rs4964287, rs4595619, rs10861201, rs11111997, rs4246270, and rs4246271) were scored in 118 DILI patients and in 120 drug-matched controls without liver injury.
RESULTS
No differences were found between the frequencies of any of the 7 SNPs in the cases and controls; however, a significant association was found between a TTA haplotype composed of rs10735393, rs4964287, and rs4595619 and DILI using an allele model (odds ratio, 1.79; 95% confidence interval, 1.18-2.73; P=0.008; Bonferroni corrected P=0.024).
CONCLUSIONS
These results suggest that genetic variations in TXNRD1 favor the development of DILI, although a larger confirmative study is needed.
Keyword
MeSH Terms
-
Alleles
Animals
Anti-Bacterial Agents
Anticonvulsants
Drug Toxicity
Drug-Induced Liver Injury
Genetic Association Studies
Genetic Variation
Haplotypes
Humans
Liver
Polymorphism, Genetic
Polymorphism, Single Nucleotide
Republic of Korea
Thioredoxin Reductase 1
Anti-Bacterial Agents
Anticonvulsants
Thioredoxin Reductase 1
Figure
Reference
-
1. Andrade RJ, Robles M, Ulzurrun E, Lucena MI. Drug-induced liver injury: insights from genetic studies. Pharmacogenomics. 2009. 10:1467–1487.2. Gunawan B, Kaplowitz N. Clinical perspectives on xenobiotic-induced hepatotoxicity. Drug Metab Rev. 2004. 36:301–312.3. Lee WM. Acute liver failure in the United States. Semin Liver Dis. 2003. 23:217–226.4. Holt MP, Ju C. Mechanisms of drug-induced liver injury. AAPS J. 2006. 8:E48–E54.5. Park BK, Kitteringham NR, Maggs JL, Pirmohamed M, Williams DP. The role of metabolic activation in drug-induced hepatotoxicity. Annu Rev Pharmacol Toxicol. 2005. 45:177–202.6. Walgren JL, Mitchell MD, Thompson DC. Role of metabolism in drug-induced idiosyncratic hepatotoxicity. Crit Rev Toxicol. 2005. 35:325–361.7. Minami K, Saito T, Narahara M, Tomita H, Kato H, Sugiyama H, Katoh M, Nakajima M, Yokoi T. Relationship between hepatic gene expression profiles and hepatotoxicity in five typical hepatotoxicant-administered rats. Toxicol Sci. 2005. 87:296–305.8. Zidek N, Hellmann J, Kramer PJ, Hewitt PG. Acute hepatotoxicity: a predictive model based on focused illumina microarrays. Toxicol Sci. 2007. 99:289–302.9. Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000. 346(Pt 1):1–8.10. Fontana RJ, Watkins PB, Bonkovsky HL, Chalasani N, Davern T, Serrano J, Rochon J. DILIN Study Group. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf. 2009. 32:55–68.11. Naranjo CA, Busto U, Sellers EM, Sandor P, Ruiz I, Roberts EA, Janecek E, Domecq C, Greenblatt DJ. A method for estimating the probability of adverse drug reactions. Clin Pharmacol Ther. 1981. 30:239–245.12. Naranjo CA, Shear NH, Lanctôt KL. Advances in the diagnosis of adverse drug reactions. J Clin Pharmacol. 1992. 32:897–904.13. Han W, Kang D, Park IA, Kim SW, Bae JY, Chung KW, Noh DY. Associations between breast cancer susceptibility gene polymorphisms and clinicopathological features. Clin Cancer Res. 2004. 10:124–130.14. Ju C, Pohl LR. Tolerogenic role of Kupffer cells in immune-mediated adverse drug reactions. Toxicology. 2005. 209:109–112.15. Choi JH, Ahn BM, Yi J, Lee JH, Nam SW, Chon CY, Han KH, Ahn SH, Jang IJ, Cho JY, Suh Y, Cho MO, Lee JE, Kim KH, Lee MG. MRP2 haplotypes confer differential susceptibility to toxic liver injury. Pharmacogenet Genomics. 2007. 17:403–415.16. Daly AK, Aithal GP, Leathart JB, Swainsbury RA, Dang TS, Day CP. Genetic susceptibility to diclofenac-induced hepatotoxicity: contribution of UGT2B7, CYP2C8, and ABCC2 genotypes. Gastroenterology. 2007. 132:272–281.17. Huang YS, Su WJ, Huang YH, Chen CY, Chang FY, Lin HC, Lee SD. Genetic polymorphisms of manganese superoxide dismutase, NAD(P)H:quinone oxidoreductase, glutathione S-transferase M1 and T1, and the susceptibility to drug-induced liver injury. J Hepatol. 2007. 47:128–134.18. Lucena MI, Andrade RJ, Martínez C, Ulzurrun E, García-Martín E, Borraz Y, Fernández MC, Romero-Gomez M, Castiella A, Planas R, Costa J, Anzola S, Agúndez JA. Spanish Group for the Study of Drug-Induced Liver Disease. Glutathione S-transferase m1 and t1 null genotypes increase susceptibility to idiosyncratic drug-induced liver injury. Hepatology. 2008. 48:588–596.19. Roy B, Chowdhury A, Kundu S, Santra A, Dey B, Chakraborty M, Majumder PP. Increased risk of antituberculosis drug-induced hepatotoxicity in individuals with glutathione S-transferase M1 'null' mutation. J Gastroenterol Hepatol. 2001. 16:1033–1037.20. Watanabe I, Tomita A, Shimizu M, Sugawara M, Yasumo H, Koishi R, Takahashi T, Miyoshi K, Nakamura K, Izumi T, Matsushita Y, Furukawa H, Haruyama H, Koga T. A study to survey susceptible genetic factors responsible for troglitazone-associated hepatotoxicity in Japanese patients with type 2 diabetes mellitus. Clin Pharmacol Ther. 2003. 73:435–455.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Do Natural Health Products Cause Toxic Hepatitis?
- Drug Induced Liver Injury by Prophylactic Administration of Albendazole
- Medicinal Herbs and Toxic Hepatitis
- Genetic polymorphisms of interleukin-10 and transforming growth factor-β1 and antituberculosis drugs-induced liver injury
- Factors affecting drug-induced liver injury: antithyroid drugs as instances