Healthc Inform Res.
2014 Oct;20(4):295-303. 10.4258/hir.2014.20.4.295.
Clinical Data Element Ontology for Unified Indexing and Retrieval of Data Elements across Multiple Metadata Registries
- Affiliations
-
- 1National Center for Medical Information and Knowledge, Korea National Institute of Health, Cheongju, Korea.
- 2Seoul National University Biomedical Informatics (SNUBI), Seoul National University College of Medicine, Seoul, Korea. juhan@snu.ac.kr
- 3Department of Biomedical Informatics, Asan Medical Center, Seoul, Korea.
- 4Systems Biomedical Informatics National Core Research Center (SBI-NCRC), Seoul National University College of Medicine, Seoul, Korea.
- KMID: 2166805
- DOI: http://doi.org/10.4258/hir.2014.20.4.295
Abstract
OBJECTIVES
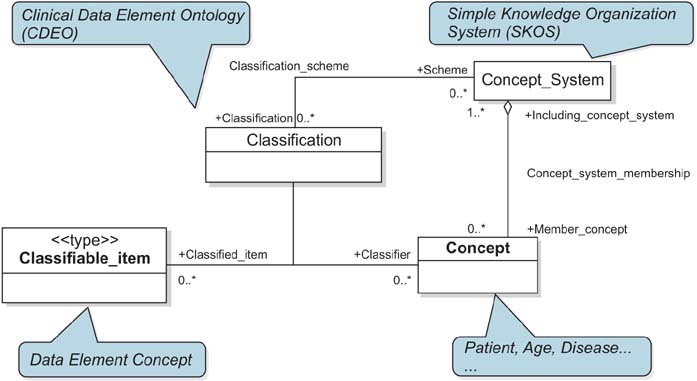
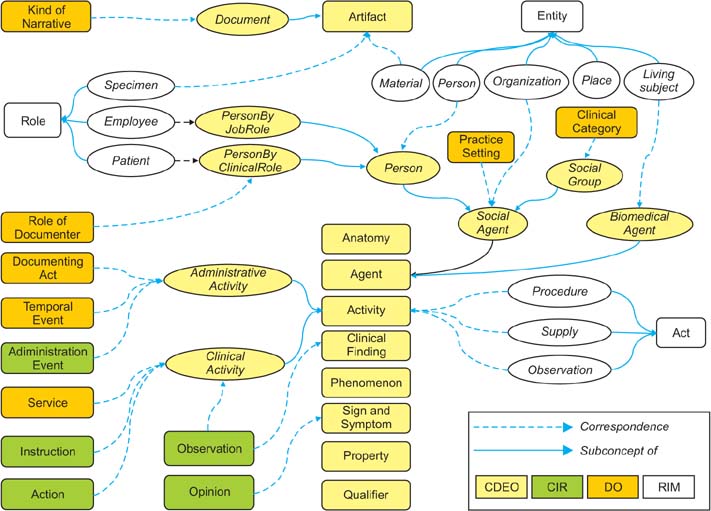
Classification of data elements (DEs), which is used in clinical documents is challenging, even in across ISO/IEC 11179 compliant clinical metadata registries (MDRs) due to no existence of reliable standard for identifying DEs. We suggest the Clinical Data Element Ontology (CDEO) for unified indexing and retrieval of DEs across MDRs.
METHODS
The CDEO was developed through harmonization of existing clinical document models and empirical analysis of MDRs. For specific classification as using data element concept (DEC), The Simple Knowledge Organization System was chosen to represent and organize the DECs. Six basic requirements also were set that the CDEO must meet, including indexing target to be a DEC, organizing DECs using their semantic relationships. For evaluation of the CDEO, three indexers mapped 400 DECs to more than 1 CDEO term in order to determine whether the CDEO produces a consistent index to a given DEC. The level of agreement among the indexers was determined by calculating the intraclass correlation coefficient (ICC).
RESULTS
We developed CDEO with 578 concepts. Through two application use-case scenarios, usability of the CDEO is evaluated and it fully met all of the considered requirements. The ICC among the three indexers was estimated to be 0.59 (95% confidence interval, 0.52-0.66).
CONCLUSIONS
The CDEO organizes DECs originating from different MDRs into a single unified conceptual structure. It enables highly selective search and retrieval of relevant DEs from multiple MDRs for clinical documentation and clinical research data aggregation.
MeSH Terms
Figure
Reference
-
1. International Organization for Standardization. Information technology: Metadata registries (MDR): Part 1. Framework. Geneva, Switzerland: International Organization for Standardization;2004. ISO/IEC 11179-1:2004(E).2. Radford MJ, Arnold JM, Bennett SJ, Cinquegrani MP, Cleland JG, Havranek EP, et al. ACC/AHA key data elements and definitions for measuring the clinical management and outcomes of patients with chronic heart failure: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Data Standards (Writing Committee to Develop Heart Failure Clinical Data Standards): developed in collaboration with the American College of Chest Physicians and the International Society for Heart and Lung Transplantation: endorsed by the Heart Failure Society of America. Circulation. 2005; 112(12):1888–1916.
Article3. Stover PJ, Harlan WR, Hammond JA, Hendershot T, Hamilton CM. PhenX: a toolkit for interdisciplinary genetics research. Curr Opin Lipidol. 2010; 21(2):136–140.
Article4. CDISC Inc. CDISC Shared Health and Research Electronic Library pilot report (version 1.0). Austin (TX): CDISC Inc.;2010.5. caBIG Strategic Planning Workspace. The Cancer Biomedical Informatics Grid (caBIG): infrastructure and applications for a worldwide research community. Stud Health Technol Inform. 2007; 129(Pt 1):330–334.6. Loring DW, Lowenstein DH, Barbaro NM, Fureman BE, Odenkirchen J, Jacobs MP, et al. Common data elements in epilepsy research: development and implementation of the NINDS epilepsy CDE project. Epilepsia. 2011; 52(6):1186–1191.
Article7. Warzel DB, Andonaydis C, McCurry B, Chilukuri R, Ishmukhamedov S, Covitz P. Common data element (CDE) management and deployment in clinical trials. AMIA Annu Symp Proc. 2003; 2003:1048.8. Tobias J, Chilukuri R, Komatsoulis GA, Mohanty S, Sioutos N, Warzel DB, et al. The CAP cancer protocols: a case study of caCORE based data standards implementation to integrate with the Cancer Biomedical Informatics Grid. BMC Med Inform Decis Mak. 2006; 6:25.9. Pathak J, Wang J, Kashyap S, Basford M, Li R, Masys DR, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc. 2011; 18(4):376–386.
Article10. Papatheodorou I, Crichton C, Morris L, Maccallum P, Davies J, et al. METABRIC Group. A metadata approach for clinical data management in translational genomics studies in breast cancer. BMC Med Genomics. 2009; 2:66.
Article11. Miller AC, Odenkirchen J, Duhaime AC, Hicks R. Common data elements for research on traumatic brain injury: pediatric considerations. J Neurotrauma. 2012; 29(4):634–638.
Article12. International Organization for Standardization. Information technology: Metadata registries (MDR): Part 2. Classification. Geneva, Switzerland: International Organization for Standardization;2005. ISO/IEC 11179-2:2005(E).13. National Cancer Institute. CDE browser [Internet]. Washington (DC): US National Institutes of Health;2011. cited at 2014 Oct 1. Available from: http://cdebrowser.nci.nih.gov/CDEBrowser/.14. Nadkarni PM, Brandt CA. The Common Data Elements for cancer research: remarks on functions and structure. Methods Inf Med. 2006; 45(6):594–601.
Article15. Australian Institute of Health and Welfare. METeOR Browse registry [Internet]. Canberra: Australian Institute of Health and Welfare;2010. cited at 2014 Oct 1. Available from: http://meteor.aihw.gov.au/content/index.phtml/itemId/237704.16. Park YR, Kim JH. Metadata registry and management system based on ISO 11179 for Cancer Clinical Trials Information System. AMIA Annu Symp Proc. 2006; 2006:1056.17. Frazier P, Rossi-Mori A, Dolin RH, Alschuler L, Huff SM. The creation of an ontology of clinical document names. Stud Health Technol Inform. 2001; 84(Pt 1):94–98.18. Shapiro JS, Bakken S, Hyun S, Melton GB, Schlegel C, Johnson SB. Document ontology: supporting narrative documents in electronic health records. AMIA Annu Symp Proc. 2005; 2005:684–688.19. Chen ES, Melton GB, Engelstad ME, Sarkar IN. Standardizing Clinical Document Names Using the HL7/LOINC Document Ontology and LOINC Codes. AMIA Annu Symp Proc. 2010; 2010:101–105.20. Kashyap V, Morales A, Hongsermeier T, Li Q. Definitions management: a semantics-based approach for clinical documentation in healthcare delivery. In : Proceedings of the 4th International Semantic Web Conference (ISWC 2005); 2005 Nov 6-10; Galway, Ireland. p. 887–901.21. Kim MK, Hwang SH, Kim HG, Kang YK, Choi HC, Kim DS. Classification and conceptualization of clinical documents using formal concept analysis. J Korean Soc Med Inform. 2006; 12(1):31–43.
Article22. Song TM, Park HA, Jin DL. Development of health information search engine based on metadata and ontology. Healthc Inform Res. 2014; 20(2):88–98.
Article23. W3C. SKOS Simple Knowledge Organization System Reference [Internet]. place unknown: World Wide Web Consortium;2009. cited at 2014 Oct 1. Available from: http://www.w3.org/TR/skos-reference/.24. International Organization for Standardization. Information technology: Metadata registries (MDR): Part 3. Registry metamodel and basic attributes. Geneva, Switzerland: International Organization for Standardization;2013. ISO/IEC 11179-3:2013(E).25. Beale T, Heard S. An ontology-based model of clinical information. Stud Health Technol Inform. 2007; 129(Pt 1):760–764.26. Health Level Seven International. HL7 version 3: Reference Information Model [Internet]. Ann Arbor (MI): Health Level Seven International;2004. cited at 2014 Oct 1. Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=77.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Development of Health Information Search Engine Based on Metadata and Ontology
- A Study of Effective Unified Medical Language System Concept Indexing in Radiology Reports
- Design of an Integration System for Bioinformatics Data Sources Using a Global MDR
- Computerization of radiology departmental management using the personal computer: application in medium sized hospital
- Biotea-2-Bioschemas, facilitating structured markup for semantically annotated scholarly publications