Healthc Inform Res.
2013 Mar;19(1):16-24. 10.4258/hir.2013.19.1.16.
Comparison of Machine Learning Algorithms for Classification of the Sentences in Three Clinical Practice Guidelines
- Affiliations
-
- 1Information and Communication Science, Semyung University, Jecheon, Korea.
- 2IT Department, Gachon University, Incheon, Korea. ugkang@gachon.ac.kr
- KMID: 2166656
- DOI: http://doi.org/10.4258/hir.2013.19.1.16
Abstract
OBJECTIVES
Clinical Practice Guidelines (CPGs) are an effective tool for minimizing the gap between a physician's clinical decision and medical evidence and for modeling the systematic and standardized pathway used to provide better medical treatment to patients.
METHODS
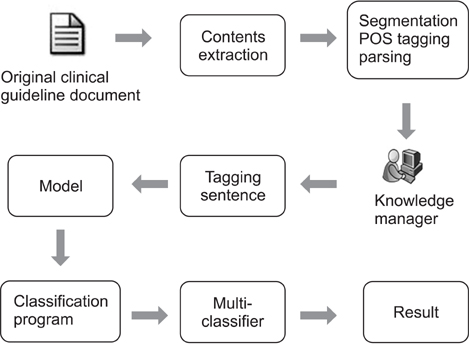
In this study, sentences within the clinical guidelines are categorized according to a classification system. We used three clinical guidelines that incorporated knowledge from medical experts in the field of family medicine. These were the seventh report of the Joint National Committee (JNC7) on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure from the National Heart, Lung, and Blood Institute; the third report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults from the same institution; and the Standards of Medical Care in Diabetes 2010 report from the American Diabetes Association. Three annotators each tagged 346 sentences hand-chosen from these three clinical guidelines. The three annotators then carried out cross-validations of the tagged corpus. We also used various machine learning-based classifiers for sentence classification.
RESULTS
We conducted experiments using real-valued features and token units, as well as a Boolean feature. The results showed that the combination of maximum entropy-based learning and information gain-based feature extraction gave the best classification performance (over 98% f-measure) in four sentence categories.
CONCLUSIONS
This result confirmed the contribution of the feature reduction algorithm and optimal technique for very sparse feature spaces, such as the sentence classification problem in the clinical guideline document.
MeSH Terms
Figure
Reference
-
1. Woolf SH, Grol R, Hutchinson A, Eccles M, Grimshaw J. Clinical guidelines: potential benefits, limitations, and harms of clinical guidelines. BMJ. 1999. 318(7182):527–530.
Article2. Buchanan BG, Shortliffe EH. Rule-based expert systems: the MYCIN experiments of the Stanford Heuristic Programming Project. 1984. Reading (MA): Addison-Wesley.3. Kim SN, Martinez D, Cavedon L, Yencken L. Automatic classification of sentences to support Evidence Based Medicine. BMC Bioinformatics. 2011. 12:Suppl 2. S5.
Article4. Nawaz R, Thompson P, McNaught J, Ananiadou S. Meta-knowledge annotation of bio-events. Proceedings of the International Conference on Language Resources and Evaluation. 2010 May 17-23; Valletta, Malta. 2498–2505.5. Pan F. Multi-dimensional fragment classification in biomedical text [dissertation]. 2006. Ottawa, Canada: Queen's University.6. Duda RO, Hart PE, Stork DG. Pattern classification. 2000. 2nd ed. New York (NY): Wiley.7. Heckerman D. Bayesian networks for data mining. Data Min Knowl Discov. 1997. 1(1):79–119.8. Tan PN, Steinbach M, Kumar V. Introduction to data mining. 2006. Boston (MA): Pearson Addison-Wesley.9. Cho SB, Won HH. Machine learning in DNA microarray analysis for cancer classification. Proceedings of the 1st Asia-Pacific Bioinformatics Conference on Bioinformatics. 2003 Feb 4-7; Adelaide, Australia. 189–198.10. Vapnik VN. Statistical learning theory. 1998. Wiley: New York (NY).11. Cristianini N, Shawe-Taylor J. An introduction to support vector machines: and other kernel-based learning methods. 2000. Cambridge (MA): Cambridge University Press.12. Liu H, Yu L. Toward integrating feature selection algorithms for classification and clustering. IEEE Trans Knowl Data Eng. 2005. 17(4):491–502.
Article13. Khan A, Baharudin B, Lee LH, Khan K. A review of machine learning algorithms for text-documents classification. J Adv Inf Technol. 2010. 1(1):4–20.14. Yang Y, Pedersen JO. A comparative study on feature selection in text categorization. Proceedings of the 14th International Conference on Machine Learning. 1997 Jul 8-12; Nashville, TN. –412. –420.15. Lee C, Lee GG. Information gain and divergence-based feature selection for machine learning-based text categorization. Inf Process Manag. 2006. 42(1):155–165.
Article16. Silla CN Jr, Pappa GL, Freitas AA, Kaestner CA. Lemaitre C, Reyes CA, Gonzalez JA, editors. Automatic text summarization with genetic algorithm-based attribute selection. Advances in Artificial Intelligence, Lecture Notes in Computer Science. 2004. vol. 3315. Heidelberg, Germany: Springer;305–314.
Article17. The National Heart, Lung, and Blood Institute. The seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. 2004. Bethesda (MD): National Institutes of Health.18. Third report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (adult treatment panel III). 2002. Bethesda (MD): National Institutes of Health.19. American Diabetes Association. Standards of medical care in diabetes: 2010. Diabetes Care. 2010. 33:Suppl 1. S11–S61.20. Shatkay H, Pan F, Rzhetsky A, Wilbur WJ. Multi-dimensional classification of biomedical text: toward automated, practical provision of high-utility text to diverse users. Bioinformatics. 2008. 24(18):2086–2093.
Article21. Thompson P, Venturi G, McNaught J, Montemagni S, Ananiadou S. Categorising modality in biomedical texts. Proceedings of the LREC 2008 Workshop on Building and Evaluating Resources for Biomedical Text Mining. 2008 May 28-30; Marrakech, Morocco. 27–34.22. Feldman R, Sanger J. The text mining handbook: advanced approaches in analyzing unstructured data. 2007. Cambridge (MA): Cambridge University Press.23. The Stanford Natural Language Processing Group. The Stanford parser: a statistical parser [Internet]. c2012. cited at 2013 Mar 18. Stanford (CA): Stanford NLP Group;Available from: http://nlp.stanford.edu/software/lex-parser.shtml.24. Song MH, Kim SH, Park DK, Lee YH. A multi-classifier based guideline sentence classification system. Healthc Inform Res. 2011. 17(4):224–231.
Article25. Machine Learning Group at University of Waikato. Weka3: data mining software in Java [Internet]. c2012. cited at 2013 Mar 18. Hamilton, New Zealand: The University of Waikato;Available from: http://www.cs.waikato.ac.nz/ml/weka/.26. Machine Learning Group at University of Waikato. Weka sources [Internet]. c2012. cited at 2013 Mar 18. Hamilton, New Zealand: The University of Waikato;Available from: http://weka.sourceforge.net/doc.stable/.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Research and Trends in Orthopedic Surgery and Musculoskeletal Tumor Using Artificial Intelligence
- Determination of the stage and grade of periodontitis according to the current classification of periodontal and peri-implant diseases and conditions (2018) using machine learning algorithms
- Artificial Intelligence for Clinical Research in Voice Disease
- Applications of Machine Learning Using Electronic Medical Records in Spine Surgery
- Artificial intelligence, machine learning, and deep learning in women’s health nursing