Healthc Inform Res.
2011 Sep;17(3):143-149. 10.4258/hir.2011.17.3.143.
The Recent Progress in Quantitative Medical Image Analysis for Computer Aided Diagnosis Systems
- Affiliations
-
- 1Biomedical Engineering Branch, National Cancer Center, Goyang, Korea. kimkg@ncc.re.kr
- KMID: 1974239
- DOI: http://doi.org/10.4258/hir.2011.17.3.143
Abstract
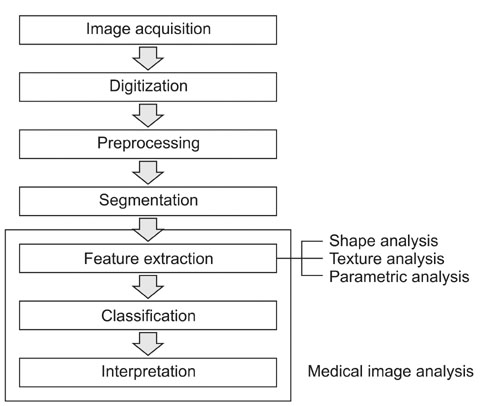
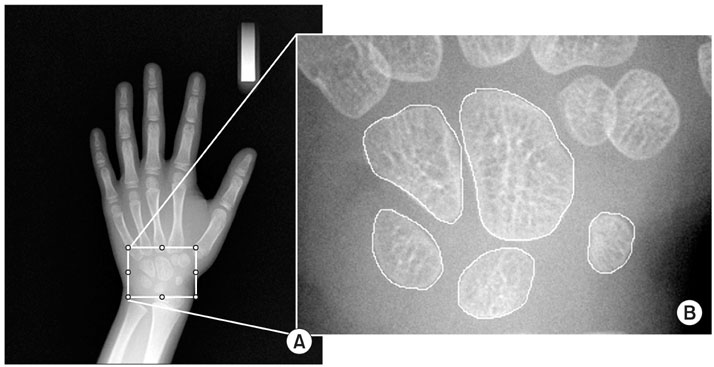
- Computer-aided diagnosis (CAD) has become one of the major research subjects in medical imaging and diagnostic radiology. Many different CAD schemes are being developed for use in the detection and/or characterization of various lesions found through various types of medical imaging. These imaging technologies employ conventional projection radiography, computed tomography, magnetic resonance imaging, ultrasonography, etc. In order to achieve a high performance level for a computerized diagnosis, it is important to employ effective image analysis techniques in the major steps of a CAD scheme. The main objective of this review is to attempt to introduce the diverse methods used for quantitative image analysis, and to provide a guide for clinicians.
Keyword
MeSH Terms
Figure
Cited by 3 articles
-
Quantitative Measurement Method for Possible Rib Fractures in Chest Radiographs
Jaeil Kim, Sungjun Kim, Young Jae Kim, Kwang Gi Kim, Jinah Park
Healthc Inform Res. 2013;19(3):196-204. doi: 10.4258/hir.2013.19.3.196.Automated Detection of Retinal Nerve Fiber Layer by Texture-Based Analysis for Glaucoma Evaluation
Anindita Septiarini, Agus Harjoko, Reza Pulungan, Retno Ekantini
Healthc Inform Res. 2018;24(4):335-345. doi: 10.4258/hir.2018.24.4.335.Automatic Glaucoma Detection Method Applying a Statistical Approach to Fundus Images
Anindita Septiarini, Dyna M. Khairina, Awang H. Kridalaksana, Hamdani Hamdani
Healthc Inform Res. 2018;24(1):53-60. doi: 10.4258/hir.2018.24.1.53.
Reference
-
1. Verma B, Zakos J. A computer-aided diagnosis system for digital mammograms based on fuzzy-neural and feature extraction techniques. IEEE Trans Inf Technol Biomed. 2001. 5:46–54.
Article2. Gletsos M, Mougiakakou SG, Matsopoulos GK, Nikita KS, Nikita AS, Kelekis D. A computer-aided diagnostic system to characterize CT focal liver lesions: design and optimization of a neural network classifier. IEEE Trans Inf Technol Biomed. 2003. 7:153–162.
Article3. Schmid-Saugeona P, Guillodb J, Thirana JP. Towards a computer-aided diagnosis system for pigmented skin lesions. Comput Med Imaging Graph. 2003. 27:65–78.
Article4. Gur D, Sumkin JH, Rockette HE, Ganott M, Hakim C, Hardesty L, Poller WR, Shah R, Wallace L. Changes in breast cancer detection and mammography recall rates after the introduction of a computer-aided detection system. J Natl Cancer Inst. 2004. 96:185–190.
Article5. Deserno TM. Biomedical image processing. 2011. Berlin: Springer Heidelberg.6. Rodenacker K, Bengtsson E. A feature set for cytometry on digitized microscopic images. Anal Cell Pathol. 2003. 25:1–36.
Article7. Pathak SD, Ng L, Wyman B, Fogarasi S, Racki S, Oelund JC, Sparks B, Chalana V. Quantitative image analysis: software systems in drug development trials. Drug Discov Today. 2003. 8:451–458.
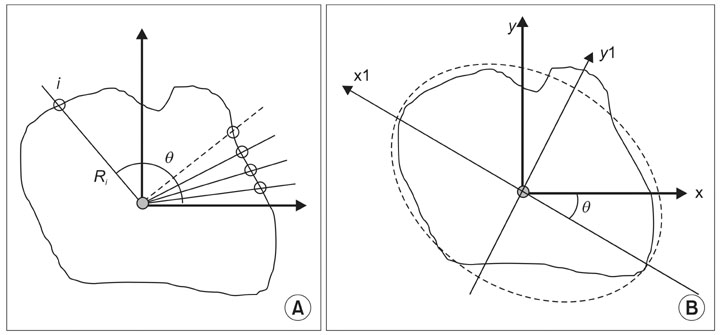
Article8. Clark MW. Quantitative shape analysis: a review. Math Geol. 1981. 13:303–320.
Article9. Rahbar G, Sie AC, Hansen GC, Prince JS, Melany ML, Reynolds HE, Jackson VP, Sayre JW, Bassett LW. Benign versus malignant solid breast masses: US differentiation. Radiology. 1999. 213:889–894.
Article10. Liberman L, Morris EA, Lee MJ, Kaplan JB, LaTrenta LR, Menell JH, Abramson AF, Dashnaw SM, Ballon DJ, Dershaw DD. Breast lesions detected on MR imaging: features and positive predictive value. AJR Am J Roentgenol. 2002. 179:171–178.
Article11. Drabycz S, Stockwell RG, Mitchell JR. Image texture characterization using the discrete orthonormal S-transform. J Digit Imaging. 2009. 22:696–708.
Article12. Ji Q, Engel J, Craine E. Classifying cervix tissue patterns with texture analysis. Pattern Recognit. 2000. 33:1561–1573.
Article13. Galloway MM. Texture analysis using gray level run lengths. Comput Graph Image Process. 1975. 4:172–179.
Article14. Cross GR, Jain AK. Markov random field texture models. IEEE Trans Pattern Anal Mach Intell. 1983. 5:25–39.
Article15. Doh SY, Park RH. Segmentation of statistical texture images using the metric space theory. Signal Process. 1996. 53:27–34.
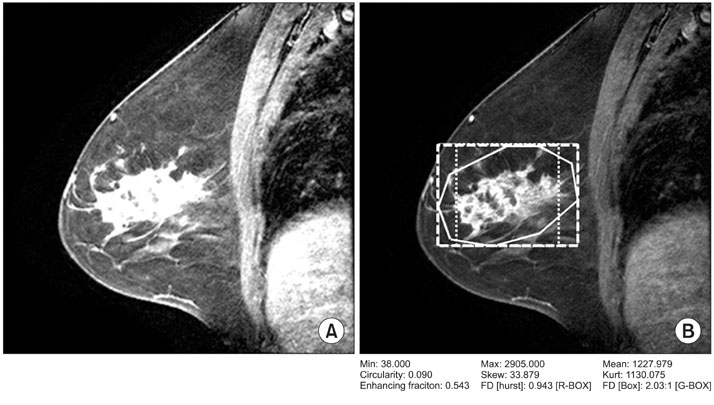
Article16. Woods BJ, Clymer BD, Kurc T, Heverhagen JT, Stevens R, Orsdemir A, Bulan O, Knopp MV. Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data. J Magn Reson Imaging. 2007. 25:495–501.
Article17. Ojala T, Pietikainen M, Kyllonen J. Co-occurrence histograms via learning vector quantization. Proceedings of the 11th Scandinavian Conference in Image Analysis. 1990. Kangerlussuaq, Greenland. 103–108.18. Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med. 2003. 50:92–98.
Article19. Arivazhagan S, Ganesan L. Texture classification using wavelet transform. Pattern Recognit Lett. 2003. 24:1513–1521.
Article20. Jian M, Guo H, Liu L. Texture image classification using visual perceptual texture features and Gabor wavelet features. J Comput. 2009. 4:763–770.
Article21. Dettori L, Semler L. A comparison of wavelet, ridgelet, and curvelet-based texture classification algorithms in computed tomography. Comput Biol Med. 2007. 37:486–498.
Article22. Karahaliou A, Skiadopoulos S, Boniatis I, Sakellaropoulos P, Likaki E, Panayiotakis G, Costaridou L. Texture analysis of tissue surrounding microcalcifications on mammograms for breast cancer diagnosis. Br J Radiol. 2007. 80:648–656.
Article23. Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol. 2008. 15:1513–1525.
Article24. Reyes-Aldasoro CC, Bhalerao A. Volumetric texture description and discriminant feature selection for MRI. Inf Process Med Imaging. 2003. 18:282–293.
Article25. Chen Z, Ning R. Breast volume denoising and noise characterization by 3D wavelet transform. Comput Med Imaging Graph. 2004. 28:235–246.
Article26. Ranguelova E, Quinn A. Analysis and synthesis of threedimensional Gaussian Markov random fields. Proceedings of International Conference on Image Processing (ICIP). 1999 Oct 24-28; Kobe, Japan. 430–434.
Article27. Di Nallo AM, Vidiri A, Marzi S, Mirri A, Fabi A, Carapella CM, Pace A, Crecco M. Quantitative analysis of CT-perfusion parameters in the evaluation of brain gliomas and metastases. J Exp Clin Cancer Res. 2009. 28:38.
Article28. Osanai O, Ohtsuka M, Hotta M, Kitaharai T, Takema Y. A new method for the visualization and quantification of internal skin elasticity by ultrasound imaging. Skin Res Technol. 2011. Feb 23 [Epub]. http://dx.doi.org/10.1111/j.1600-0846.2010.00492.x.
Article29. Johnson RA, Wichern DW. Applied multi-variate statistical analysis. 2002. 5th ed. Englewood Cliffs, NJ: Prentice-Hall.30. Lu D, Weng Q. A survey of image classification methods and techniques for improving classification performance. Int J Remote Sens. 2007. 28:823–870.
Article31. Gose E, Johnsonbaugh R, Jost S. Pattern recognition and image analysis. 1996. Englewood Cliffs, NJ: Prentice-Hall.32. Li S, Kwok JT, Zhu H, Wang Y. Texture classification using the support vector machines. Pattern Recognit. 2003. 36:2883–2893.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Development of Intelligent Picture Archiving Communication System that Embedded Computer-Aided Diagnosis and Speech Recognition
- Computer-Aided Diagnosis in Chest CT
- An Engineering View on Megatrends in Radiology: Digitization to Quantitative Tools of Medicine
- A Computer-aided Design Tool with Semiautomatic Image-Processing Features for Visualizing Biological Pathways
- Use of the surface-based registration function of computer-aided design/computer-aided manufacturing software in medical simulation software for three-dimensional simulation of orthognathic surgery