J Korean Med Sci.
2007 Jun;22(3):425-430. 10.3346/jkms.2007.22.3.425.
Distinct Linkage Disequilibrium (LD) Runs of Single Nucleotide Polymorphisms and Microsatellite Markers; Implications for Use of Mixed Marker Haplotypes in LD-based Mapping
- Affiliations
-
- 1Department of Laboratory Medicine, College of Medicine, Hallym University, Chunchon, Korea.
- 2Department of Laboratory Medicine, College of Medicine, Yonsei University, Seoul, Korea.
- 3Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. jwonk@smc.samsung.co.kr
- KMID: 1778348
- DOI: http://doi.org/10.3346/jkms.2007.22.3.425
Abstract
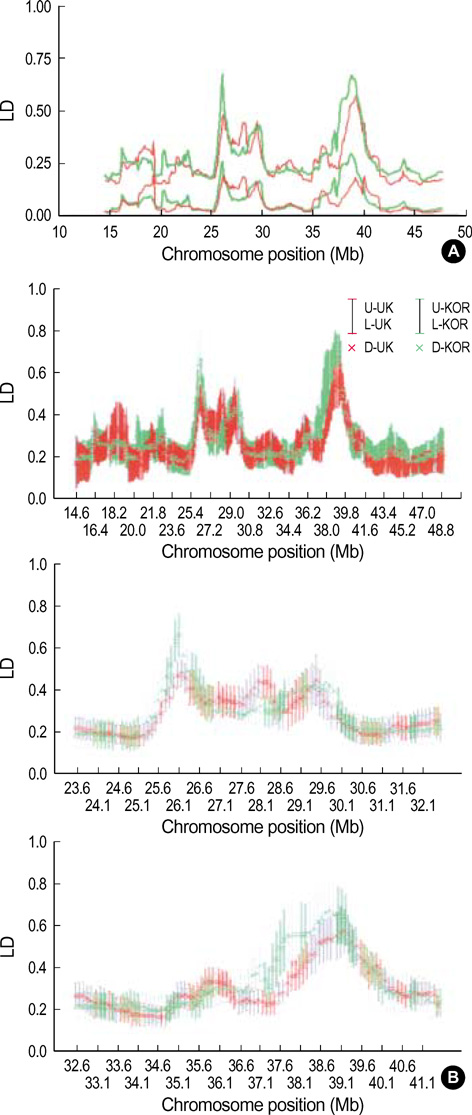
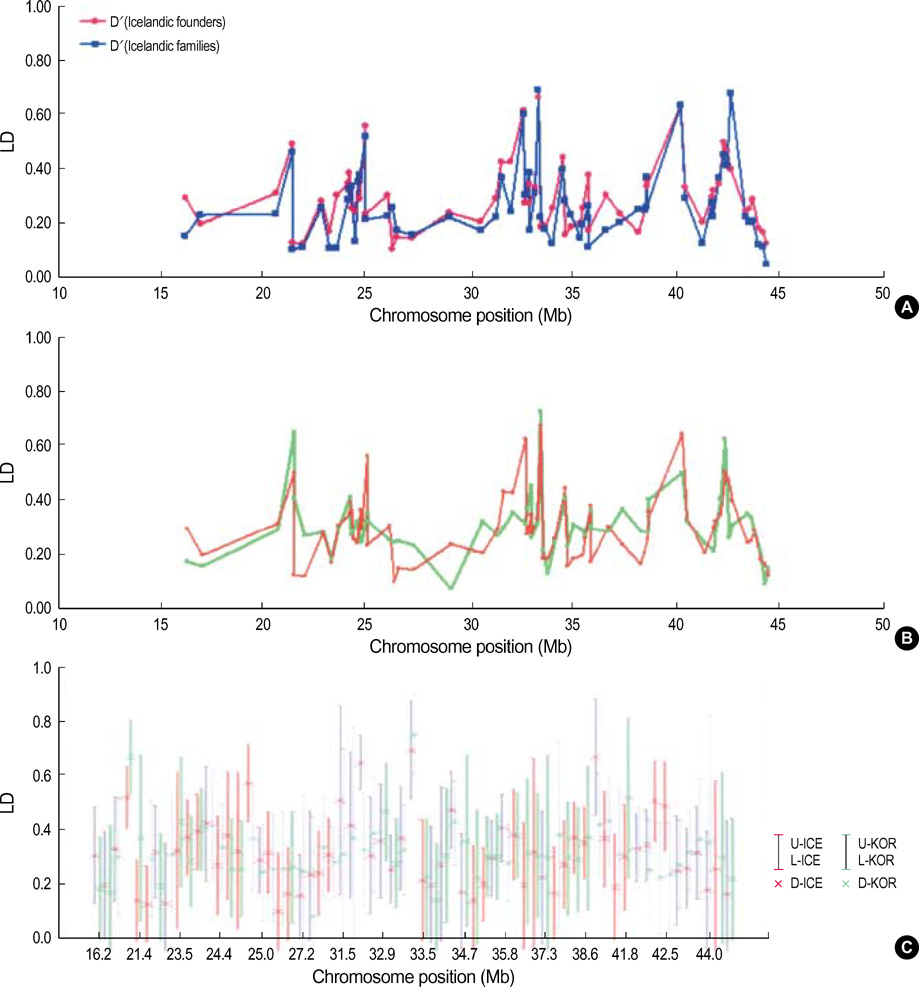
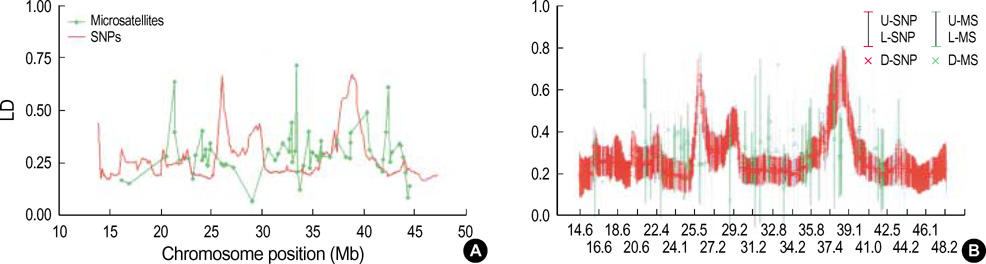
- It has been suggested that the haplotypic relationship between microsatellite markers and single nucleotide polymorphisms (SNPs) is of considerable importance, as microsatellite markers can potentially be incorporated into haplotypes containing SNPs to increase marker density across a region of interest. However, SNPs and microsatellite markers have different mutation rates and durations, and it is conceivable that the linkage disequilibrium (LD) patterns between the genetic markers may considerably differ. We assessed the LD patterns using 1,661 SNPs and 65 microsatellite markers along chromosome 22 and investigated whether common patterns of LD between the two genetic markers are deduced from the results. The results demonstrated that the patterns of LD among microsatellite markers varied considerably and the LD runs of SNPs and microsatellite markers showed distinct patterns. Microsatellite markers have a much higher mutation rate and the evolution of microsatellite markers is a more complex process which has distinct mutation properties from those of SNPs. We consider that these might contribute to the different LD patterns between the two genetic markers. Therefore, it would seem inadvisable to make assumptions about persistence of LD across even a relatively small genetic distance among microsatellite markers and to construct mixed marker haplotypes/LD maps employing microsatellite markers.
MeSH Terms
Figure
Reference
-
1. Mohlke KL, Lange EM, Valle TT, Ghosh S, Magnuson VL, Silander K, Watanabe RM, Chines PS, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. Linkage disequilibrium between microsatellite markers extends beyond 1 cM on chromosome 20 in Finns. Genome Res. 2001. 11:1221–1226.
Article2. Merriman TR, Eaves IA, Twells RC, Merriman ME, Danoy PA, Muxworthy CE, Hunter KM, Cox RD, Cucca F, McKinney PA, Shield JP, Baum JD, Tuomilehto J, Tuomilehto-Wolf E, Ionesco-Tirgoviste C, Joner G, Thorsby E, Undlien DE, Pociot F, Nerup J, Ronningen KS, Bain SC, Todd JA. Transmission of haplotypes of microsatellite markers rather than single marker alleles in the mapping of a putative type 1 diabetes susceptibility gene (IDDM6). Hum Mol Genet. 1998. 7:517–524.3. Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R Jr, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996. 13:399–408.
Article4. Burgner D, Rockett K, Ackerman H, Hull J, Usen S, Pinder M, Kwiatkowski DP. Haplotypic relationship between SNP and microsatellite markers at the NOS2A locus in two populations. Genes Immun. 2003. 4:506–514.
Article5. Nakajima T, Jorde LB, Ishigami T, Umemura S, Emi M, Lalouel JM, Inoue I. Nucleotide diversity and haplotype structure of the human angiotensinogen gene in two populations. Am J Hum Genet. 2002. 70:108–123.
Article6. Varilo T, Paunio T, Parker A, Perola M, Meyer J, Terwilliger JD, Peltonen L. The interval of linkage disequilibrium (LD) detected with microsatellite and SNP markers in chromosomes of Finnish populations with different histories. Hum Mol Genet. 2003. 1:51–59.
Article7. Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996. 271:1380–1387.
Article8. Tishkoff SA, Goldman A, Calafell F, Speed WC, Deinard AS, Bonne-Tamir B, Kidd JR, Pakstis AJ, Jenkins T, Kidd KK. A global haplotype analysis of the myotonic dystrophy locus: implications for the evolution of modern humans and for the origin of myotonic dystrophy mutations. Am J Hum Genet. 1998. 62:1389–1402.
Article9. Kidd KK, Morar B, Castiglione CM, Zhao H, Pakstis AJ, Speed WC, Bonne-Tamir B, Lu RB, Goldman D, Lee C, Nam YS, Grandy DK, Jenkins T, Kidd JR. A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet. 1998. 103:211–227.
Article10. Tishkoff SA, Pakstis AJ, Stoneking M, Kidd JR, Destro-Bisol G, Sanjantila A, Lu RB, Deinard AS, Sirugo G, Jenkins T, Kidd KK, Clark AG. Short tandem-repeat polymorphism/alu haplotype variation at the PLAT locus: implications for modern human origins. Am J Hum Genet. 2000. 67:901–925.
Article11. Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, Dibling T, Tinsley E, Kirby S, Carter D, Papaspyridonos M, Livingstone S, Ganske R, Lohmussaar E, Zernant J, Tonisson N, Remm M, Magi R, Puurand T, Vilo J, Kurg A, Rice K, Deloukas P, Mott R, Metspalu A, Bentley DR, Cardon LR, Dunham I. A first-generation linkage disequilibrium map of human chromosome 22. Nature. 2002. 1:544–548.
Article12. Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005. 307:1072–1079.
Article13. Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, Nguyen BT, Norris MC, Sheehan JB, Shen N, Stern D, Stokowski RP, Thomas DJ, Trulson MO, Vyas KR, Frazer KA, Fodor SP, Cox DR. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001. 294:1719–1723.
Article14. Zavattari P, Deidda E, Whalen M, Lampis R, Mulargia A, Loddo M, Eaves I, Mastio G, Todd JA, Cucca F. Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: demography, chromosome recombination frequency and selection. Hum Mol Genet. 2000. 12:2947–2957.
Article15. Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002. 31:241–247.
Article16. Aulchenko YS, Axenovich TI, Mackay I, van Duijn CM. miLD and booLD programs for calculation and analysis of corrected linkage disequilibrium. Ann Hum Genet. 2003. 67:372–375.
Article17. Brookes AJ. The essence of SNPs. Gene. 1999. 234:177–186.
Article18. Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004. 5:435–445.
Article19. Kruglyak S, Durrett RT, Schug MD, Aquadro CF. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc Natl Acad Sci USA. 1998. 95:10774–10778.
Article20. Santibanez-Koref MF, Gangeswaran R, Hancock JM. A relationship between lengths of microsatellites and nearby substitution rates in mammalian genomes. Mol Biol Evol. 2001. 18:2119–2123.21. Amos W, Sawcer SJ, Feakes RW, Rubinsztein DC. Microsatellites show mutational bias and heterozygote instability. Nat Genet. 1996. 13:390–391.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- An Alternative Way of Constructing Ancestral Graphs Using Marker Allele Ages from Population Linkage Disequilibrium Information
- Short Reads Phasing to Construct Haplotypes in Genomic Regions That Are Associated with Body Mass Index in Korean Individuals
- Controlling Linkage Disequilibrium in Association Tests: Revisiting APOE Association in Alzheimer's Disease
- Characterization of Single Nucleotide Polymorphisms in 55 Disease-Associated Genes in a Korean Population
- Effects of Single Nucleotide Polymorphism Marker Density on Haplotype Block Partition