Korean J Orthod.
2008 Apr;38(2):133-143. 10.4041/kjod.2008.38.2.133.
Characteristics of MSX1 gene in Korean nonsyndromic cleft lip and palate individuals
- Affiliations
-
- 1Department of Orthodontics, School of Dentistry, Pusan National University, Korea. softid@pusan.ac.kr
- KMID: 1481500
- DOI: http://doi.org/10.4041/kjod.2008.38.2.133
Abstract
OBJECTIVE
This study was performed to identify the characteristics of the MSX1 gene (locus chromosome 4p16) in Korean nonsyndromic cleft lip and palate (CL/P), which is assumed to be a major candidate gene acting as a causal factor in nonsyndromic CL/P and missing teeth.
METHODS
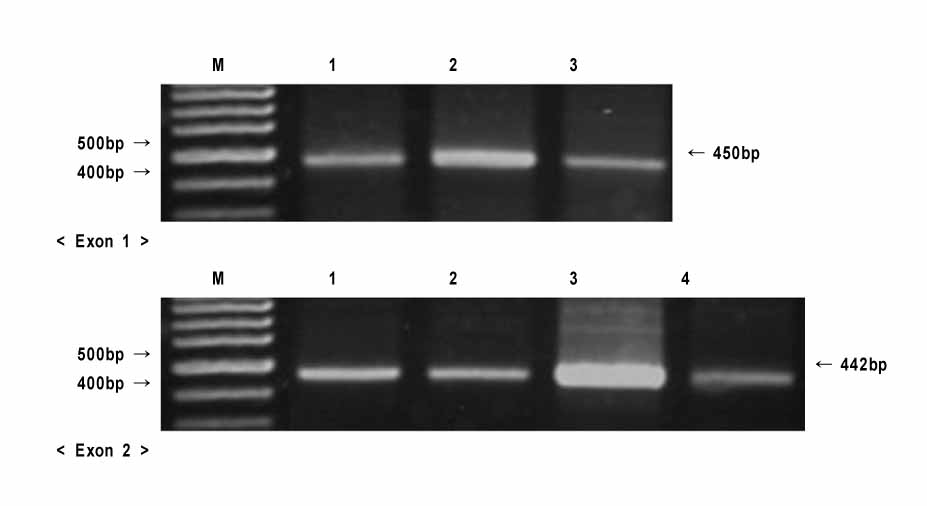
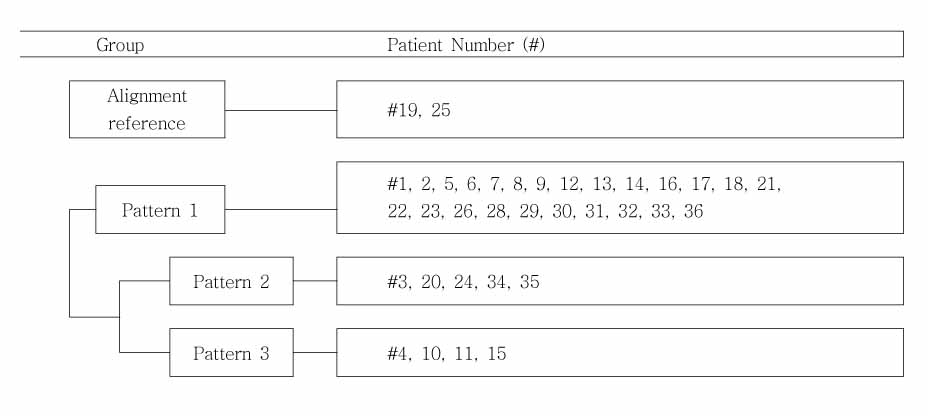
The 36 individuals (23 males and 13 females) who had visited the department of orthodontics at from 1998 to 2002 and who had nonsyndromic CL/P were included in the study. Using a PCR-based assay, the MSX1 gene was amplified, sequenced, and searched for inferred protein products (Reference: Homo sapiens MSX1, accession number AF426432 and NP_002439). The common single nucleotide polymorphisms were observed.
RESULTS
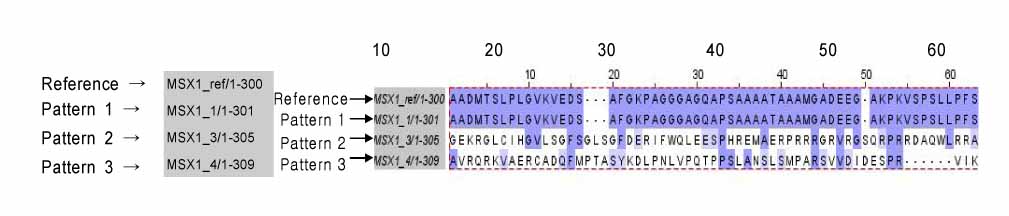
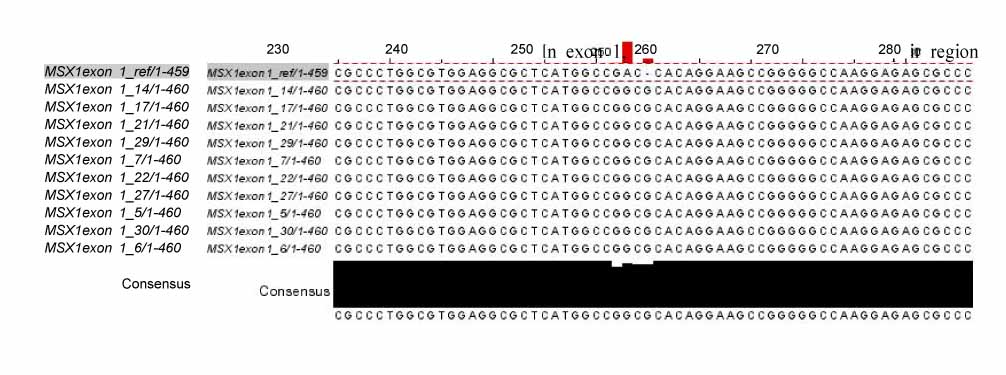
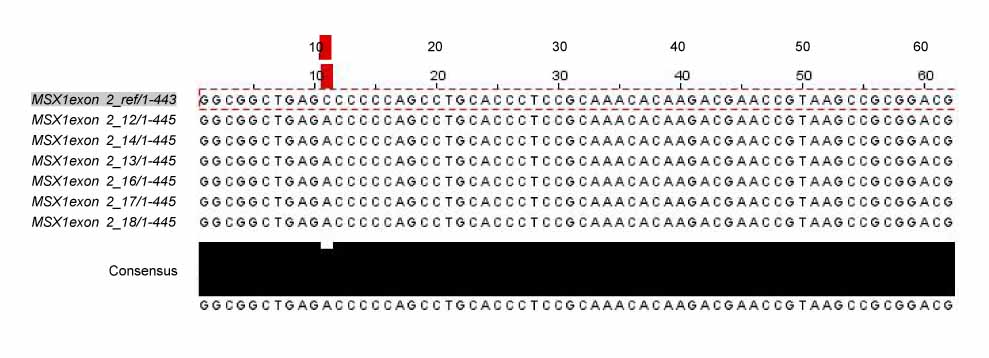
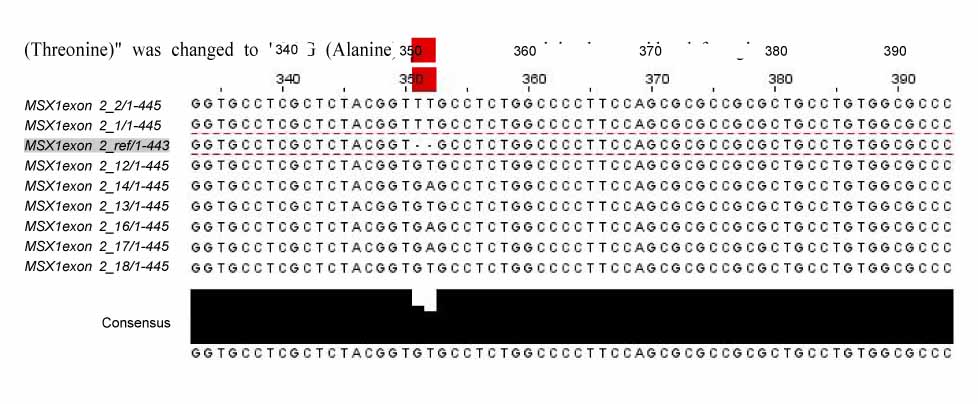
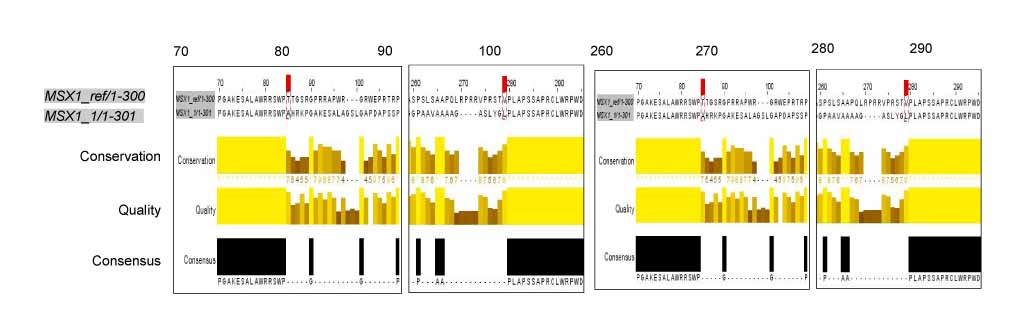
In exon 1, nucleotide "A" of the 253 basepair (bp) region was substituted for "G", and in the 255 bp region, nucleotide "G" was inserted. In exon 2, nucleotide "C" of the 11 bp region was substituted for "A", and "T" or "G" was inserted into the 351 bp region whereas "T" or "A" was inserted into the 352 bp region. In protein analysis, "Thr85Ala" missense mutation was found. The "Thr85Ala" missense mutation in this study is different from those of studies using subjects of other races.
CONCLUSIONS
The results suggest that there is specific mutation of MSX1 in Korean and it plays an important role in Korean nonsyndromic CL/P. However, any distinct genetic polymorphisms between CL/P with missing teeth in the cleft region and CL/P without missing teeth could not be found.
MeSH Terms
Figure
Reference
-
1. Croen LA, Shaw GM, Wasserman CR, Tolarova MM. Racial and ethnic variations in the prevalence of orofacial clefts in California, 1983-1992. Am J Med Genet. 1998. 79:42–47.
Article2. Vanderas AP. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft palate J. 1987. 24:216–225.3. Lidral AC, Murray JC, Buetow KH, Basart AM, Schearer H, Shiang R, et al. Studies of the candidate genes TGFB2, MSX1, TGFA, and TGFB3 in the etiology of cleft lip and palate in the Philippines. Cleft Palate Craniofac J. 1997. 34:1–6.
Article4. Schutte BC, Murray JC. The many faces and factors of orofacial clefts. Hum Mol Genet. 1999. 8:1853–1859.5. Kim S, Kim WJ, Oh C, Kim JC. Cleft lip and palate incidence among the live births in the republic of Korea. J Korean Med Sci. 2002. 17:49–52.
Article6. Hecht JT, Wang Y, Blanton SH, Daiger SP. Van der Woude syndrome and nonsyndromic cleft lip and palate. Am J Hum Genet. 1992. 51:442–444.7. Schutte BC, Bjork BC, Coppage KB, Malik MI, Gregory SG, Scott DJ, et al. A preliminary gene map for the Van der Woude syndrome critical region derived from 900 kb of genomic sequence at 1q32-q41. Genome Res. 2000. 10:81–94.8. Murray JC. Face facts: genes, environment, and clefts. Am J Hum Genet. 1995. 57:227–232.9. Lander ES, Schork N. Genetic dissection of complex traits. Science. 1994. 265:2037–2048.
Article10. Hecht JT, Yang P, Michels VV, Buetow KH. Complex segregation analysis of nonsyndromic cleft lip and palate. Am J Hum Genet. 1991. 49:674–681.11. Wyszynski DF, Beaty TH, Maestri NE. Genetics of nonsyndromic oral clefts revisited. Cleft Palate Craniofac J. 1996. 33:406–417.
Article12. Jones MC. Etiology of prospective evaluation of 428 patients. Cleft Palate J. 1988. 25:16–20.13. Murray JC. Gene/environment causes of cleft lip and palate. Clin Genet. 2002. 61:248–256.14. Kim SS, Son WS. Sequencing analysis of the OFC1 gene on the nonsyndromic cleft lip and palate patient in Korean. Korean J Orthod. 2003. 33:185–197.15. Murray JC, Schutte BC. Cleft palate: players, pathways, and pursuits. J Clin Invest. 2004. 113:1676–1678.
Article16. Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, et al. Association of MSX1 and TGFB3 with nonsymdromic clefting in human. Am J Hum Genet. 1998. 63:557–568.17. Jugessur A, Lie RT, Wilcox AJ, Murray JC, Taylor JA, Saugstad OD, et al. Variants of developmental genes (TGFA, TGFB3, and MSX1) and their associations with craniofacial clefts: a case-parent triad analysis. Genet Epidemiol. 2003. 24:230–239.
Article18. Satokata I, Maas R. MSX1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994. 6:348–356.
Article19. Blanco R, Chakraborty R, Barton SA, Carreno H, Paredes M, Jara L, et al. Evidence of a sex-dependent association between the MSX1 locus and nonsyndromic cleft lip with or without cleft palate in the Chilean population. Hum Biol. 2001. 73:81–89.
Article20. Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet. 2003. 40:399–407.
Article21. van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000. 24:342–343.
Article22. Slayton RL, Williams L, Murray JC, Wheeler JJ, Lidral AC, Nishimura CJ. Genetic association studies of cleft lip and/or palate with hypodontia outside the cleft region. Cleft Palate Craniofac J. 2003. 40:274–279.
Article23. Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996. 13:417–421.
Article24. Lidral AC, Reising BC. The role of MSX1 in human tooth agenesis. J Dent Res. 2002. 81:274–278.
Article25. De Muynck S, Schollen E, Matthijs G, Verdonck A, Devriendt K, Carels C. A novel MSX1 mutation in hypodontia. Am J Med Genet A. 2004. 128:401–403.26. Mattheeuws N, Dermaut L, Martens G. Has hypodontia increased in Caucasians during the 20th century? A meta-analysis. Eur J Orthod. 2004. 26:99–103.
Article27. Graber LW. Congenital absence of teeth: a review with emphasis on inheritance patterns. J Am Dent Assoc. 1978. 96:266–275.
Article28. Tsai TP, Huang CS, Huang CC, See LC. Distribution patterns of primary and permanent dentition in children with unilateral complete cleft lip and palate. Cleft palate Craniofac J. 1998. 35:154–160.
Article29. Shapira Y, Lubit E, Kuftinec MM. Congentially missing second premolars in cleft lip and cleft palate children. Am J Orthod Dentofacial Orthop. 1999. 115:396–400.
Article30. Shapira Y, Lubit E, Kuftinec MM. Hypodontia in children with various types of clefts. Angle Orthod. 2000. 70:16–21.31. Lourenco Ribeiro L, Teixeira Das Neves L, Costa B, Ribeiro Gomide M. Dental anomalies of the permanent lateral incisors and prevalence of hypodontia outside the cleft area in complete unilateral cleft lip and palate. Cleft palate Craniofac J. 2003. 40:172–175.
Article32. Olin WH. Dental anomalies in cleft lip and cleft palate patients. Angle Orthod. 1964. 34:119–123.33. Eerens K, Vlietinck R, Heidbuchel K, Van Olmen A, Derom C, Willems G, Carels C. Hypodontia and tooth formation in groups of children with cleft, siblings without cleft, and nonrelated controls. Cleft palate Craniofac J. 2001. 38:374–378.
Article34. Jumlongras D, Bei M, Stimson JM, Wang WF, DePalma SR, Seidman CE, et al. A nonsense mutation in MSX1 causes Witkop syndrome. Am J Hum Genet. 2001. 69:67–74.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Association between MSX1 SNPs and Nonsyndromic Cleft Lip with or without Cleft Palate in the Korean Population
- A cephalometric study on the position of the hyoid bone in cleft lip and palate individuals
- Genetic Analysis of TGFA, MTHFR, and IFR6 in Korean Patients Affected by Nonsyndromic Cleft Lip with or without Cleft Palate (CL/P)
- A study on the maxillary dental arch and palate of unilateral cleft lip and palate individuals
- Orthodontic consideration of cleft lip and palate (Report 1)