Ann Lab Med.
2012 Sep;32(5):370-374. 10.3343/alm.2012.32.5.370.
First Korean Case of Robinsoniella peoriensis Bacteremia in a Patient with Aspiration Pneumonia
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Hospital, Seoul, Korea.
- 2Department of Laboratory Medicine, Seoul National University College of Medicine, Seoul, Korea. m91w95pf@snu.ac.kr
- 3Department of Laboratory Medicine, Seoul National University Bundang Hospital, Seongnam, Korea.
- 4Department of Internal Medicine, Seoul National University Bundang Hospital, Seongnam, Korea.
- KMID: 1387354
- DOI: http://doi.org/10.3343/alm.2012.32.5.370
Abstract
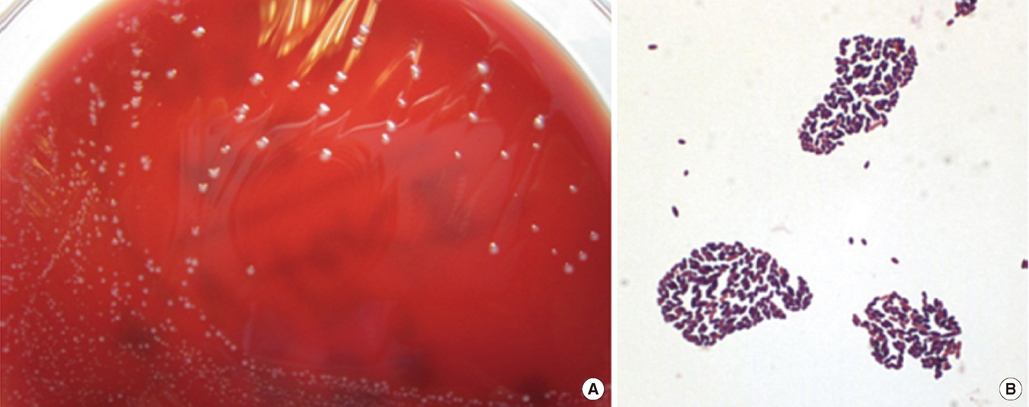
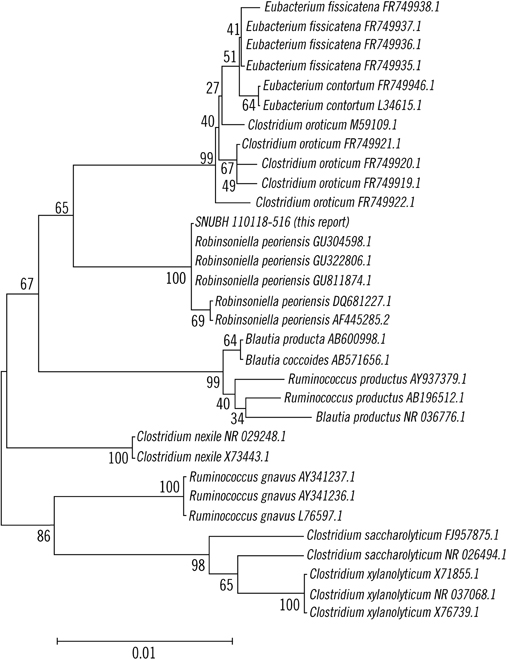
- Robinsoniella peoriensis has recently been identified as a Gram-positive, spore-forming, anaerobic rod originally recovered from swine manure storage pits. To date, 6 cases of R. peoriensis infection have been reported, including 2 cases of bacteremia, 1 of abdominal fluid collection, and 3 of wound infection. In the present study, we report a 76-yr-old man with R. peoriensis bacteremia who developed aspiration pneumonia. Gram staining of a purified colony revealed Gram-positive, rod-shaped bacteria. Biochemical identification using API 20 A (bioMerieux, France) indicated presence of Clostridium spp. We performed both 500-bp and full-gene sequencing of 16S rRNA of the isolate. The sequence was analyzed with MicroSeq ID 16S rRNA Library v2.0 (Applied Biosystems, USA), GenBank Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/genbank), and EzTaxon database v2.1 (http://www.eztaxon.org). The 500-bp 16S rRNA sequence of the blood culture isolate showed 99.16-99.79% similarity with R. peoriensis and the full-gene 16S rRNA sequence showed 98.87-99.50% similarity with R. peoriensis. The organism was confirmed as R. peoriensis by using all of the mentioned databases except for MicroSeq, which did not include the RNA sequence of this bacterium. This case suggests that identification of R. peoriensis might be challenging in clinical laboratories with no access to molecular methods, as certain commercial identification systems may not identify, or may misidentify, this organism. To the best of our knowledge, this is the first report of the isolation of R. peoriensis in Korea.
MeSH Terms
Figure
Cited by 1 articles
-
Robinsoniella peoriensis Bacteremia: a Second Case in Korea
Sangeun Lim, Hee Jae Huh, Nam Yong Lee, Eun-Jeong Joo, Joon-Sup Yeom, Seungjun Lee, Hee-Yeon Woo, Hyosoon Park, Min-Jung Kwon
Ann Lab Med. 2017;37(4):349-351. doi: 10.3343/alm.2017.37.4.349.
Reference
-
1. Finegold SM. The role of anaerobes in human infections. Scand J Infect Dis Suppl. 1981. 26:9–13.2. Kedzia A, Kwapisz E, Wierzbowska M. Incidence of anaerobic bacteria in respiratory tract infections. Pneumonol Alergol Pol. 2003. 71:68–73.3. Cotta MA, Whitehead TR, Falsen E, Moore E, Lawson PA. Robinsoniella peoriensis gen. nov., sp. nov., isolated from a swine-manure storage pit and a human clinical source. Int J Syst Evol Microbiol. 2009. 59:150–155.4. Shen D, Chen R, Ye L, Luo Y, Tang YW. Robinsoniella peoriensis bacteremia in a patient with pancreatic cancer. J Clin Microbiol. 2010. 48:3448–3450.5. Gomez E, Gustafson DR, Colgrove R, Ly T, Santana R, Rosenblatt JE, et al. Isolation of Robinsoniella peoriensis from four human specimens. J Clin Microbiol. 2011. 49:458–460.6. Hall L, Doerr KA, Wohlfiel SL, Roberts GD. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J Clin Microbiol. 2003. 41:1447–1453.
Article7. Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007. 57:2259–2261.
Article8. Devulder G, Perrière G, Baty F, Flandrois JP. BIBI, a bioinformatics bacterial identification tool. J Clin Microbiol. 2003. 41:1785–1787.
Article9. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007. 24:1596–1599.
Article10. Cotta MA, Whitehead TR, Zeltwanger RL. Isolation, characterization and comparison of bacteria from swine faeces and manure storage pits. Environ Microbiol. 2003. 5:737–745.
Article11. Brook I, Fraizer EH. Role of anaerobic bacteria in liver abscesses in children. Pediatr Infect Dis J. 1993. 12:743–747.
Article12. Edmiston CE Jr, Krepel CJ, Seabrook GR, Jochimsen WG. Anaerobic infections in the surgical patient: microbial etiology and therapy. Clin Infect Dis. 2002. 35:S112–S118.
Article13. Dennis LS, Amy EB, et al. Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, editors. Clostridium. Manual of clinical microbiology. 2011. 10th ed. Washington, DC: ASM Press;834–857.14. Lau SK, Ng KH, Woo PC, Yip KT, Fung AM, Woo GK, et al. Usefulness of the MicroSeq 500 16S rDNA bacterial identification system for identification of anaerobic Gram positive bacilli isolated from blood cultures. J Clin Pathol. 2006. 59:219–222.
Article15. Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, Leung KW, et al. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J Clin Microbiol. 2003. 41:1996–2001.
Article16. Fontana C, Favaro M, Pelliccioni M, Pistoia ES, Favalli C. Use of the MicroSeq 500 16S rRNA gene-based sequencing for identification of bacterial isolates that commercial automated systems failed to identify correctly. J Clin Microbiol. 2005. 43:615–619.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Robinsoniella peoriensis Bacteremia: a Second Case in Korea
- Predictive value of procalcitonin for bacteremia in patients with pneumonia in the emergency department
- Aspiration pneumonia in the child with DiGeorge syndrome: A case report
- Pneumonia due to aspiration of povidine iodine after induction of general anesthesia -A case report-
- Pneumonitis and pneumonia after aspiration