Korean J Lab Med.
2011 Jan;31(1):49-53. 10.3343/kjlm.2011.31.1.49.
Miller-Dieker Syndrome with der(17)t(12;17)(q24.33;p13.3)pat Presenting with a Potential Risk of Mis-identification as a de novo Submicroscopic Deletion of 17p13.3
- Affiliations
-
- 1Department of Laboratory Medicine, Pusan National University Hospital, Busan, Korea. mindcatch@hanmail.net
- 2Department of Pediatrics, Pusan National University Yangsan Hospital, Yangsan, Korea.
- 3Department of Rehabilitation Medicine, Pusan National University Hospital, Busan, Korea.
- 4Green Cross Reference Laboratory, Yongin, Korea.
- 5Medical Research Institute, Pusan National University Hospital, Busan, Korea.
- KMID: 1096661
- DOI: http://doi.org/10.3343/kjlm.2011.31.1.49
Abstract
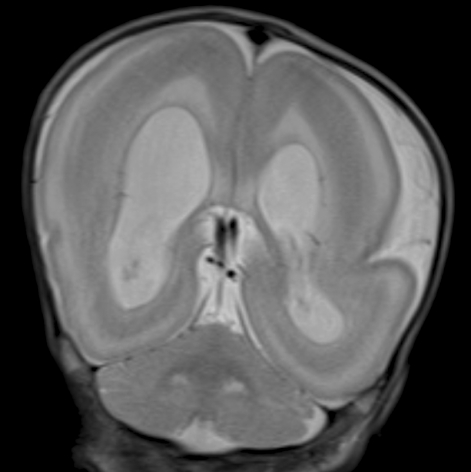
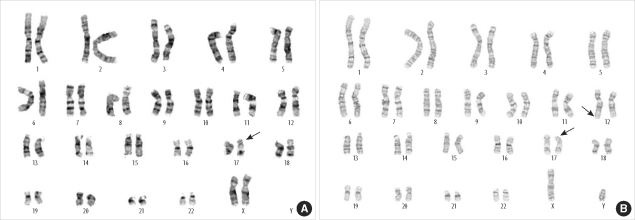
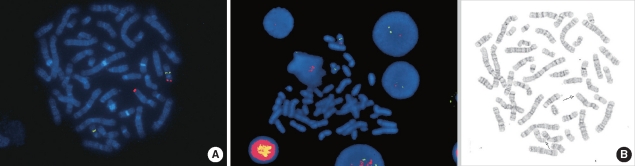
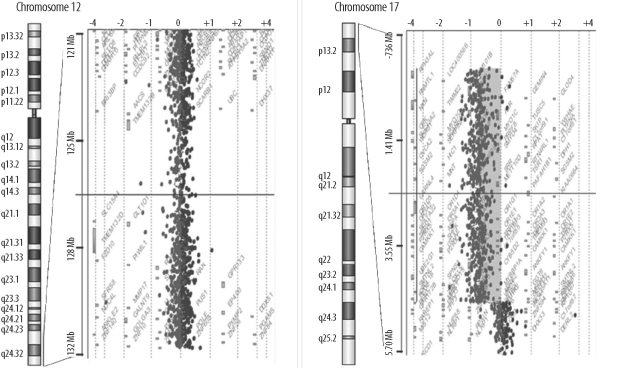
- Miller-Dieker syndrome involves a severe type of lissencephaly, which is caused by defects in the lissencephaly gene (LIS1). We report the case of a female infant with der(17)t(12;17)(q24.33;p13.3)pat caused by an unbalanced segregation of the parental balanced translocation of 17p with other chromosomes. The proband presented with facial dysmorphism, arthrogryposis, and intrauterine growth retardation. Most cases of Miller-Dieker syndrome have a de novo deletion involving 17p13.3. When Miller-Dieker syndrome is caused by an unbalanced translocation, mild-to-severe phenotypes occur according to the extension of the involved partner chromosome. However, a pure partial monosomy derived from a paternal balanced translocation is relatively rare. In this case, the submicroscopic cryptic deletion in the proband was initially elucidated by FISH, and karyotype analysis did not reveal additional chromosome abnormalities such as translocation. However, a family history of recurrent pregnancy abnormalities strongly suggested familial translocation. Sequential G-banding and FISH analysis of the father's chromosomes showed that the segment of 17p13.3-->pter was attached to the 12qter. Thus, we report a case that showed resemblance to the findings in cases of a nearly pure 17p deletion, derived from t(12;17), and delineated by whole genome array comparative genomic hybridization (CGH). If such cases are incorrectly diagnosed as Miller-Dieker syndrome caused by de novo 17p13.3 deletion, the resultant improper genetic counseling may make it difficult to exactly predict the potential risk of recurrent lissencephaly for successive pregnancies.
Keyword
MeSH Terms
-
Abnormalities, Multiple/genetics
Adult
Brain/abnormalities
Chromosome Banding
Chromosome Segregation
*Chromosomes, Human, Pair 12
*Chromosomes, Human, Pair 17
Classical Lissencephalies and Subcortical Band Heterotopias/*diagnosis
Female
Gene Deletion
Humans
In Situ Hybridization, Fluorescence
Infant, Newborn
Karyotyping
Magnetic Resonance Imaging
Male
Phenotype
Risk
Translocation, Genetic
Figure
Reference
-
1. Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993; 364:717–721. PMID: 8355785.2. Schwartz CE, Johnson JP, Holycross B, Mandeville TM, Sears TS, Graul EA, et al. Detection of submicroscopic deletions in band 17p13 in patients with the Miller-Dieker syndrome. Am J Hum Genet. 1988; 43:597–604. PMID: 2903661.3. Sharief N, Craze J, Summers D, Butler L, Wood CB. Miller-Dieker syndrome with ring chromosome 17. Arch Dis Child. 1991; 66:710–712. PMID: 1711306.
Article4. van Zelderen-Bhola SL, Breslau-Siderius EJ, Beverstock GC, Stolte-Dijkstra I, de Vries LS, Stoutenbeek P, et al. Prenatal and postnatal investigation of a case with Miller-Dieker syndrome due to a familial cryptic translocation t(17;20) (p13.3;q13.3) detected by fluorescence in situ hybridization. Prenat Diagn. 1997; 17:173–179. PMID: 9061768.5. Guerrini R, Filippi T. Neuronal migration disorders, genetics, and epileptogenesis. J Child Neurol. 2005; 20:287–299. PMID: 15921228.6. Thomas MA, Duncan AM, Bardin C, Kaloustian VM. Lissencephaly with der(17)t(17;20)(p13.3;p12.2)mat. Am J Med Genet A. 2004; 124A:292–295. PMID: 14708103.
Article7. Grosso S, Fichera M, Galesi O, Luciano D, Pucci L, Giardini F, et al. Bilateral periventricular nodular heterotopia and lissencephaly in an infant with unbalanced t(12;17)(q24.31; p13.3) translocation. Dev Med Child Neurol. 2008; 50:473–476. PMID: 18384621.
Article8. Cardoso C, Leventer RJ, Ward HL, Toyo-Oka K, Chung J, Gross A, et al. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet. 2003; 72:918–930. PMID: 12621583.
Article9. Dobyns WB, Curry CJ, Hoyme HE, Turlington L, Ledbetter DH. Clinical and molecular diagnosis of Miller-Dieker syndrome. Am J Hum Genet. 1991; 48:584–594. PMID: 1671808.10. Mignon-Ravix C, Cacciagli P, El-Waly B, Moncla A, Milh M, Girard N, et al. Deletion of YWHAE in a patient with periventricular heterotopias and pronounced corpus callosum hypoplasia. J Med Genet. 2010; 47:132–136. PMID: 19635726.
Article11. Bao L, Schorry EK. A girl with partial trisomy 12q24.31 inherited from her father and a possible novel syndrome transmitted from her mother. Am J Med Genet A. 2005; 138:361–364. PMID: 16222678.
Article12. Schoumans J, Sanner G, Nordenskjold M, Anderlid BM. Detailed clinical description of four patients with 1.3 and 2.1 Mb chromosome imbalances derived from a familial t(12;17)(q24.33;q25.3). Am J Med Genet A. 2005; 134:254–258. PMID: 15704131.
Article13. Sathanoori M, Hu J, Murthy V, Byrnes A, Vockley J, Safier R, et al. Cryptic duplication of 12q24.33→qter in a child with Angelman syndrome-simultaneous occurrence of two unrelated cytogenetic events. Am J Med Genet A. 2007; 143A:985–994. PMID: 17394213.
Article14. Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL. Resolving the resolution of array CGH. Genomics. 2007; 89:647–653. PMID: 17276656.
Article15. Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010; 86:749–764. PMID: 20466091.
Article16. Hochstenbach R, van Binsbergen E, Engelen J, Nieuwint A, Polstra A, Poddighe P, et al. Array analysis and karyotyping: workflow consequences based on a retrospective study of 36,325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet. 2009; 52:161–169. PMID: 19362174.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Miller-Dieker Syndrome
- A Case of Miller-Dieker Syndrome with Infantile Spasm and Lennox-Gastaut Syndrome
- A Case of Miller-Dieker Syndrome without Characteristic Facial Anomaly
- A Case Report of Miller-Dieker Syndrome
- Comparing Two Diagnostic Laboratory Tests for Several Microdeletions Causing Mental Retardation Syndromes: Multiplex Ligation-Dependent Amplification vs Fluorescent In Situ Hybridization