Endocrinol Metab.
2024 Feb;39(1):176-185. 10.3803/EnM.2023.1739.
Prediction of Cardiovascular Complication in Patients with Newly Diagnosed Type 2 Diabetes Using an XGBoost/GRU-ODE-Bayes-Based Machine-Learning Algorithm
- Affiliations
-
- 1Division of Endocrinology and Metabolism, Department of Internal Medicine, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 2NAVER CLOVA AI Lab, Seongnam, Korea
- 3Department of Medical Informatics, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 4Department of Biomedicine and Health Sciences, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 5Health Promotion Center, Seoul St. Mary’s Hospital, Seoul, Korea
- KMID: 2552808
- DOI: http://doi.org/10.3803/EnM.2023.1739
Abstract
- Background
Cardiovascular disease is life-threatening yet preventable for patients with type 2 diabetes mellitus (T2DM). Because each patient with T2DM has a different risk of developing cardiovascular complications, the accurate stratification of cardiovascular risk is critical. In this study, we proposed cardiovascular risk engines based on machine-learning algorithms for newly diagnosed T2DM patients in Korea.
Methods
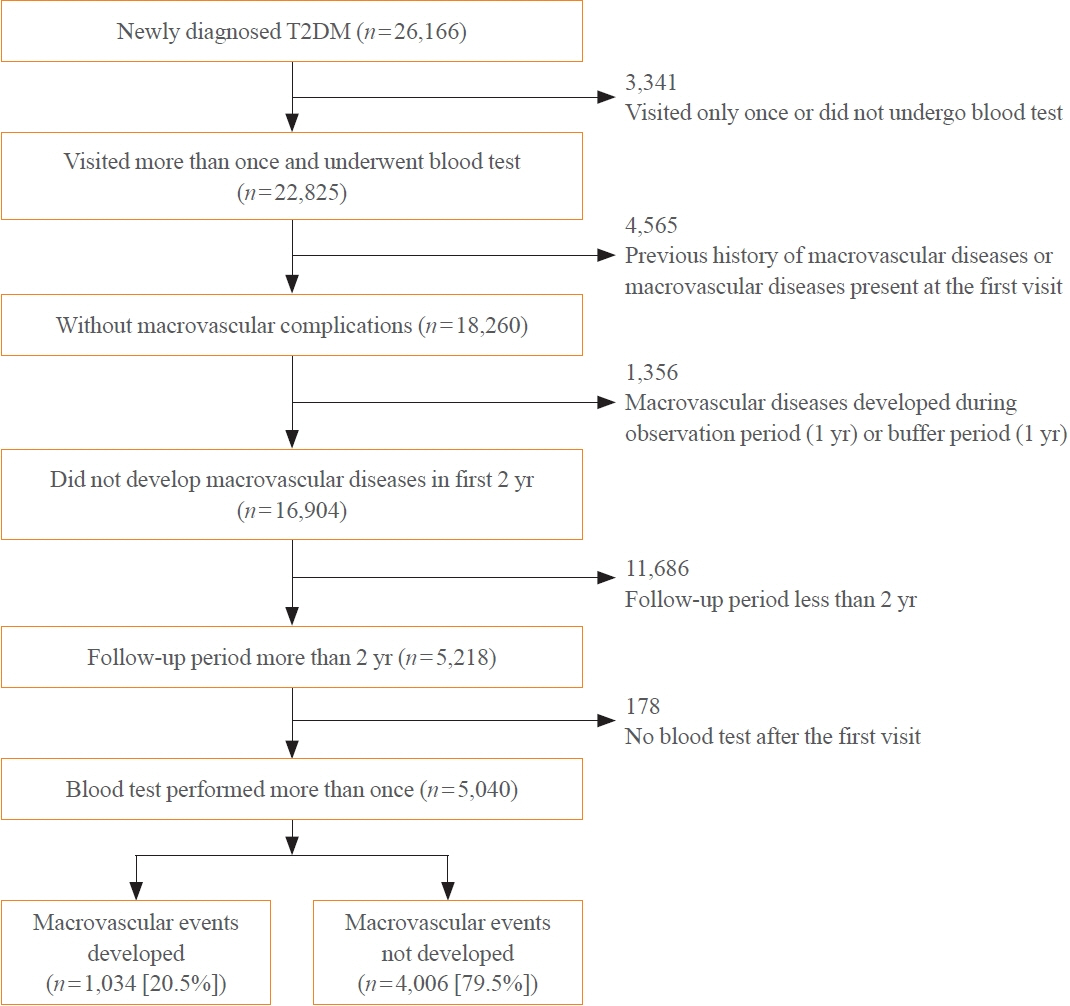
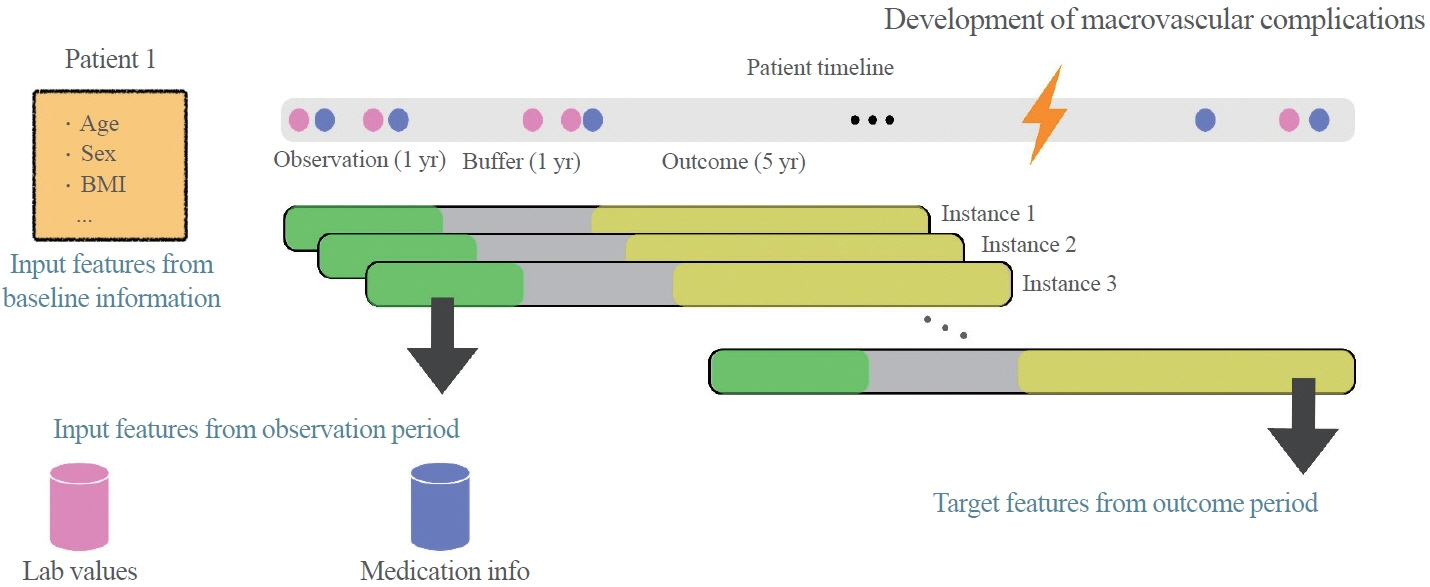
To develop the machine-learning-based cardiovascular disease engines, we retrospectively analyzed 26,166 newly diagnosed T2DM patients who visited Seoul St. Mary’s Hospital between July 2009 and April 2019. To accurately measure diabetes-related cardiovascular events, we designed a buffer (1 year), an observation (1 year), and an outcome period (5 years). The entire dataset was split into training and testing sets in an 8:2 ratio, and this procedure was repeated 100 times. The area under the receiver operating characteristic curve (AUROC) was calculated by 10-fold cross-validation on the training dataset.
Results
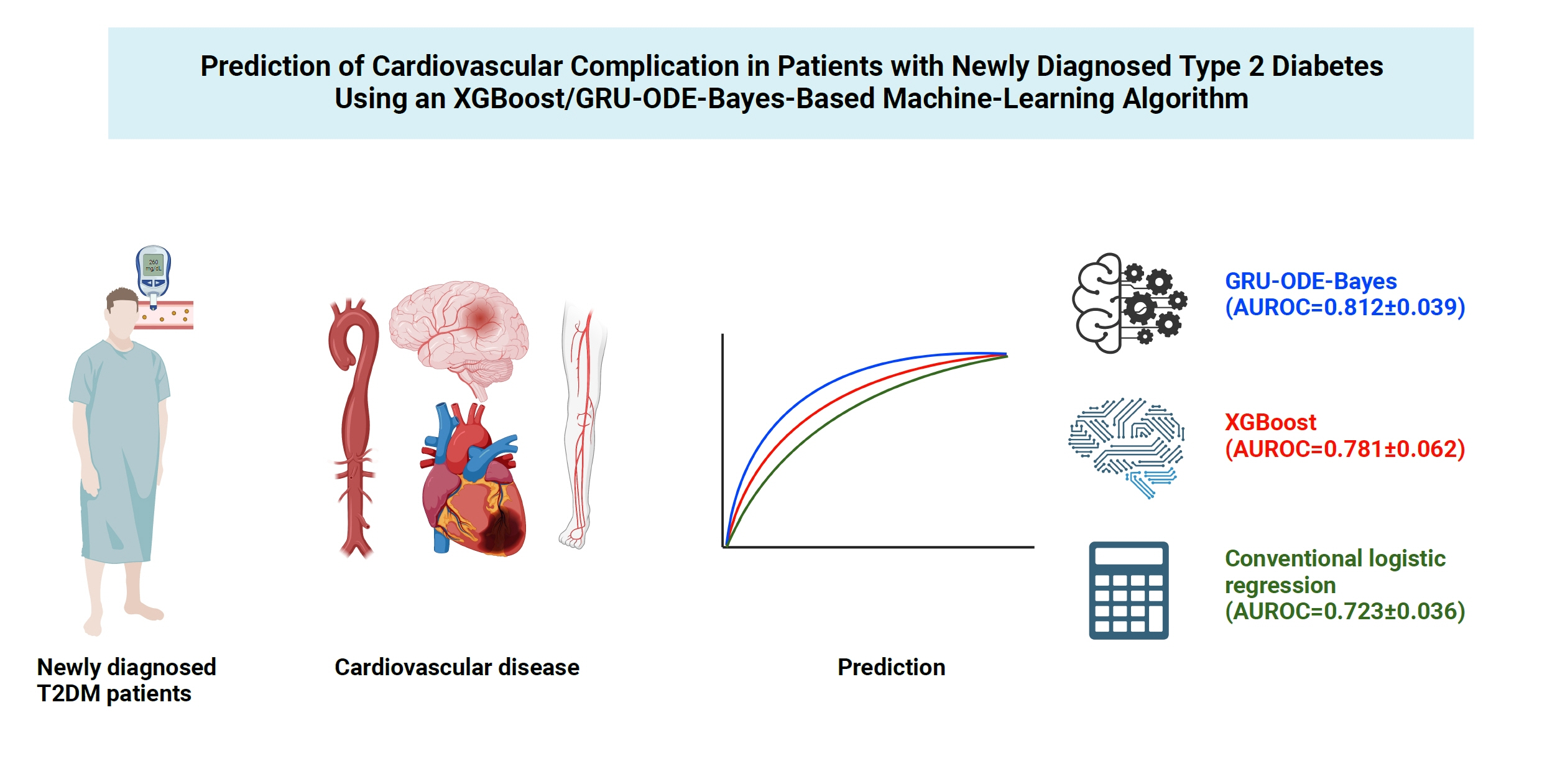
The machine-learning-based risk engines (AUROC XGBoost=0.781±0.014 and AUROC gated recurrent unit [GRU]-ordinary differential equation [ODE]-Bayes=0.812±0.016) outperformed the conventional regression-based model (AUROC=0.723± 0.036).
Conclusion
GRU-ODE-Bayes-based cardiovascular risk engine is highly accurate, easily applicable, and can provide valuable information for the individualized treatment of Korean patients with newly diagnosed T2DM.
Figure
Reference
-
1. Stratton IM, Adler AI, Neil HA, Matthews DR, Manley SE, Cull CA, et al. Association of glycaemia with macrovascular and microvascular complications of type 2 diabetes (UKPDS 35): prospective observational study. BMJ. 2000; 321:405–12.2. Emerging Risk Factors Collaboration, Sarwar N, Gao P, Seshasai SR, Gobin R, Kaptoge S, et al. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet. 2010; 375:2215–22.3. Gregg EW, Sattar N, Ali MK. The changing face of diabetes complications. Lancet Diabetes Endocrinol. 2016; 4:537–47.4. Gaede P, Lund-Andersen H, Parving HH, Pedersen O. Effect of a multifactorial intervention on mortality in type 2 diabetes. N Engl J Med. 2008; 358:580–91.5. UK Prospective Diabetes Study Group. Tight blood pressure control and risk of macrovascular and microvascular complications in type 2 diabetes: UKPDS 38. BMJ. 1998; 317:703–13.6. Pan A, Wang Y, Talaei M, Hu FB. Relation of smoking with total mortality and cardiovascular events among patients with diabetes mellitus: a meta-analysis and systematic review. Circulation. 2015; 132:1795–804.7. Colhoun HM, Betteridge DJ, Durrington PN, Hitman GA, Neil HA, Livingstone SJ, et al. Primary prevention of cardiovascular disease with atorvastatin in type 2 diabetes in the Collaborative Atorvastatin Diabetes Study (CARDS): multicentre randomised placebo-controlled trial. Lancet. 2004; 364:685–96.8. Ahlqvist E, Storm P, Karajamaki A, Martinell M, Dorkhan M, Carlsson A, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018; 6:361–9.9. Goff DC Jr, Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB Sr, Gibbons R, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association task force on practice guidelines. J Am Coll Cardiol. 2014; 63(25 Pt B):2935–59.10. Stevens RJ, Kothari V, Adler AI, Stratton IM; United Kingdom Prospective Diabetes Study (UKPDS) Group. The UKPDS risk engine: a model for the risk of coronary heart disease in type II diabetes (UKPDS 56). Clin Sci (Lond). 2001; 101:671–9.11. Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998; 97:1837–47.12. Guzder RN, Gatling W, Mullee MA, Mehta RL, Byrne CD. Prognostic value of the Framingham cardiovascular risk equation and the UKPDS risk engine for coronary heart disease in newly diagnosed type 2 diabetes: results from a United Kingdom study. Diabet Med. 2005; 22:554–62.13. van der Heijden AA, Ortegon MM, Niessen LW, Nijpels G, Dekker JM. Prediction of coronary heart disease risk in a general, pre-diabetic, and diabetic population during 10 years of follow-up: accuracy of the Framingham, SCORE, and UKPDS risk functions: the Hoorn Study. Diabetes Care. 2009; 32:2094–8.14. Yang F, Ye J, Pomerantz K, Stewart M. Potential modification of the UKPDS risk engine and evaluation of macrovascular event rates in controlled clinical trials. Diabetes Metab Syndr Obes. 2013; 6:247–56.15. Kengne AP, Patel A, Colagiuri S, Heller S, Hamet P, Marre M, et al. The Framingham and UK Prospective Diabetes Study (UKPDS) risk equations do not reliably estimate the probability of cardiovascular events in a large ethnically diverse sample of patients with diabetes: the Action in Diabetes and Vascular Disease: Preterax and Diamicron-MR Controlled Evaluation (ADVANCE) Study. Diabetologia. 2010; 53:821–31.16. McEwan P, Williams JE, Griffiths JD, Bagust A, Peters JR, Hopkinson P, et al. Evaluating the performance of the Framingham risk equations in a population with diabetes. Diabet Med. 2004; 21:318–23.17. DeFilippis AP, Young R, Carrubba CJ, McEvoy JW, Budoff MJ, Blumenthal RS, et al. An analysis of calibration and discrimination among multiple cardiovascular risk scores in a modern multiethnic cohort. Ann Intern Med. 2015; 162:266–75.18. Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018; 319:1317–8.19. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2016 Aug 13-17; San Francisco, CA. New York: Association for Computing Machinery; 2016. p. 785-94.20. Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001; 29:1189–232.21. De Brouwer E, Simm J, Arany A, Moreau Y. GRU-ODEBayes: continuous modeling of sporadically-observed time series. In: Wallach H, Larochelle H, Beygelzimer A, d’AlcheBuc F, Fox E, Garnett R. Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019); 2019 Dec 8-14; Vancouver. San Diego: Neural Information Processing Systems Foundation, Inc. (NeurIPS); 2020. p. 7347-58.22. Alaa AM, Bolton T, Di Angelantonio E, Rudd JH, van der Schaar M. Cardiovascular disease risk prediction using automated machine learning: a prospective study of 423,604 UK Biobank participants. PLoS One. 2019; 14:e0213653.23. Longato E, Fadini GP, Sparacino G, Avogaro A, Tramontan L, Di Camillo B. A deep learning approach to predict diabetes’ cardiovascular complications from administrative claims. IEEE J Biomed Health Inform. 2021; 25:3608–17.24. Ravaut M, Sadeghi H, Leung KK, Volkovs M, Kornas K, Harish V, et al. Predicting adverse outcomes due to diabetes complications with machine learning using administrative health data. NPJ Digit Med. 2021; 4:24.25. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988; 44:837–45.26. Shin SY, Kim HS. Data pseudonymization in a range that does not affect data quality: correlation with the degree of participation of clinicians. J Korean Med Sci. 2021; 36:e299.27. Kim HS, Kim DJ, Yoon KH. Medical big data is not yet available: why we need realism rather than exaggeration. Endocrinol Metab (Seoul). 2019; 34:349–54.28. Kyoung DS, Kim HS. Understanding and utilizing claim data from the Korean National Health Insurance Service (NHIS) and Health Insurance Review & Assessment (HIRA) database for research. J Lipid Atheroscler. 2022; 11:103–10.29. Lee S, Kim HS. Prospect of artificial intelligence based on electronic medical record. J Lipid Atheroscler. 2021; 10:282–90.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Machine Learning vs. Statistical Model for Prediction Modelling: Application in Medical Imaging Research
- Machine Learning Prediction for the Recurrence After Electrical Cardioversion of Patients With Persistent Atrial Fibrillation
- Statistics and Deep Belief Network-Based Cardiovascular Risk Prediction
- Understanding Arteriosclerotic Heart Disease Patients Using Electronic Health Records: A Machine Learning and Shapley Additive exPlanations Approach
- Machine Learning for the Prediction of New-Onset Diabetes Mellitus during 5-Year Follow-up in Non-Diabetic Patients with Cardiovascular Risks