Nutr Res Pract.
2023 Dec;17(6):1128-1142. 10.4162/nrp.2023.17.6.1128.
Raw Inonotus obliquus polysaccharide counteracts Alzheimer’s disease in a transgenic mouse model by activating the ubiquitin-proteosome system
- Affiliations
-
- 1School of Basic Medicine, Dali University, Dali 671000, China
- 2Department of Ophthalmology, The First Affiliated Hospital of Dali University, Dali 671000, China
- 3College of Clinical Medicine, Dali University, Dali 671000, China
- KMID: 2548869
- DOI: http://doi.org/10.4162/nrp.2023.17.6.1128
Abstract
- BACKGROUND/OBJECTIVES
Inonotus obliquus has been used as antidiabetic herb around the world, especially in the Russian and Scandinavian countries. Diabetes is widely believed to be a key factor in Alzheimer’s disease (AD), which is widely considered to be type III diabetes. To investigate whether I. obliquus can also ameliorate AD, it would be interesting to identify new clues for AD treatment. We tested the anti-AD effects of raw Inonotus obliquus polysaccharide (IOP) in a mouse model of AD (3×Tg-AD transgenic mice).
MATERIALS/METHODS
SPF-grade 3×Tg-AD mice were randomly divided into three groups (Control, Metformin, and raw IOP groups, n = 5 per group). β-Amyloid deposition in the brain was analyzed using immunohistochemistry for AD characterization. Gene and protein expression of pertinent factors of the ubiquitin-proteasome system (UPS) was determined using real-time quantitative polymerase chain reaction and Western blotting.
RESULTS
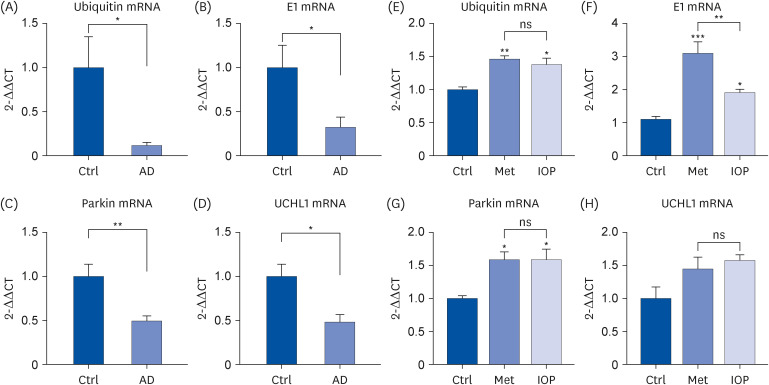
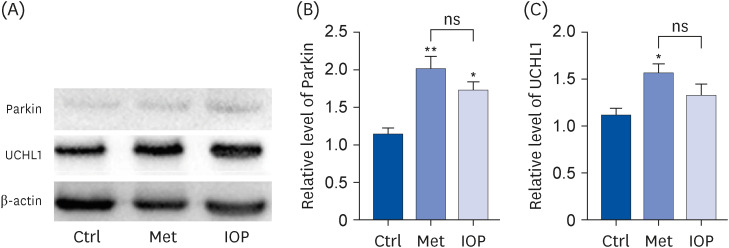
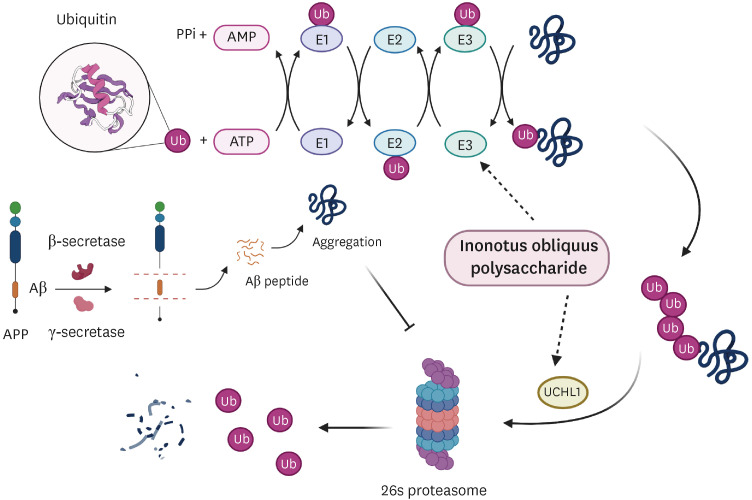
Raw IOP significantly reduced the accumulation of amyloid aggregates and facilitated UPS activity, resulting in a significant reduction in AD-related symptoms in an AD mouse model. The presence of raw IOP significantly enhanced the expression of ubiquitin, E1, and Parkin (E3) at both the mRNA and protein levels in the mouse hippocampus. The mRNA level of ubiquitin carboxyl-terminal hydrolase isozyme L1, a key factor involved in UPS activation, also increased by approximately 50%.
CONCLUSIONS
Raw IOP could contribute to AD amelioration via the UPS pathway, which could be considered as a new potential strategy for AD treatment, although we could not exclude other mechanisms involved in counteracting AD processing.
Figure
Reference
-
1. Rusek M, Pluta R, Ułamek-Kozioł M, Czuczwar SJ. Ketogenic diet in Alzheimer’s disease. Int J Mol Sci. 2019; 20:3892. PMID: 31405021.2. Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984; 120:885–890. PMID: 6375662.3. Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016; 8:595–608. PMID: 27025652.4. Cheignon C, Tomas M, Bonnefont-Rousselot D, Faller P, Hureau C, Collin F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Biol. 2018; 14:450–464. PMID: 29080524.5. Bachiller S, Alonso-Bellido IM, Real LM, Pérez-Villegas EM, Venero JL, Deierborg T, Armengol JÁ, Ruiz R. The ubiquitin proteasome system in neuromuscular disorders: moving beyond movement. Int J Mol Sci. 2020; 21:6429. PMID: 32899400.6. Hong L, Huang HC, Jiang ZF. Relationship between amyloid-beta and the ubiquitin-proteasome system in Alzheimer’s disease. Neurol Res. 2014; 36:276–282. PMID: 24512022.7. Lopez Salon M, Pasquini L, Besio Moreno M, Pasquini JM, Soto E. Relationship between beta-amyloid degradation and the 26S proteasome in neural cells. Exp Neurol. 2003; 180:131–143. PMID: 12684027.8. Verheijen BM, Stevens JA, Gentier RJ, van ’t Hekke CD, van den Hove DL, Hermes DJ, Steinbusch HW, Ruijter JM, Grimm MO, Haupenthal VJ, et al. Paradoxical effects of mutant ubiquitin on Aβ plaque formation in an Alzheimer mouse model. Neurobiol Aging. 2018; 72:62–71. PMID: 30216939.9. Lee MW, Hur H, Chang KC, Lee TS, Ka KH, Jankovsky L. Introduction to distribution and ecology of sterile conks of Inonotus obliquus . Mycobiology. 2008; 36:199–202. PMID: 23997626.10. Lu Y, Jia Y, Xue Z, Li N, Liu J, Chen H. Recent developments in Inonotus obliquus (Chaga mushroom) polysaccharides: isolation, structural characteristics, biological activities and application. Polymers (Basel). 2021; 13:1441. PMID: 33947037.11. Arata S, Watanabe J, Maeda M, Yamamoto M, Matsuhashi H, Mochizuki M, Kagami N, Honda K, Inagaki M. Continuous intake of the Chaga mushroom (Inonotus obliquus) aqueous extract suppresses cancer progression and maintains body temperature in mice. Heliyon (Lond). 2016; 2:e00111.12. Giridharan VV, Thandavarayan RA, Konishi T. Amelioration of scopolamine induced cognitive dysfunction and oxidative stress by Inonotus obliquus - a medicinal mushroom. Food Funct. 2011; 2:320–327. PMID: 21779570.13. Han Y, Nan S, Fan J, Chen Q, Zhang Y. Inonotus obliquus polysaccharides protect against Alzheimer’s disease by regulating Nrf2 signaling and exerting antioxidative and antiapoptotic effects. Int J Biol Macromol. 2019; 131:769–778. PMID: 30878614.14. Jeong JH, Kim SH, Park MN, Park JY, Park HY, Song CE, Moon JH, Choi A, Kim KD, Lee NS, et al. Water extract of mixed mushroom mycelia grown on a solid barley medium is protective against experimental focal cerebral ischemia. Curr Issues Mol Biol. 2021; 43:365–383. PMID: 34203617.15. Kou RW, Han R, Gao YQ, Li D, Yin X, Gao JM. Anti-neuroinflammatory polyoxygenated lanostanoids from Chaga mushroom Inonotus obliquus . Phytochemistry. 2021; 184:112647. PMID: 33434790.16. Lee MG, Kwon YS, Nam KS, Kim SY, Hwang IH, Kim S, Jang H. Chaga mushroom extract induces autophagy via the AMPK-mTOR signaling pathway in breast cancer cells. J Ethnopharmacol. 2021; 274:114081. PMID: 33798660.17. Li Y, Zhou Y, Wu J, Li J, Yao H. Phelligridin D from Inonotus obliquus attenuates oxidative stress and accumulation of ECM in mesangial cells under high glucose via activating Nrf2. J Nat Med. 2021; 75:1021–1029. PMID: 34052991.18. Szychowski KA, Skóra B, Pomianek T, Gmiński J. Inonotus obliquus - from folk medicine to clinical use. J Tradit Complement Med. 2020; 11:293–302. PMID: 34195023.19. Zou CX, Dong SH, Hou ZL, Yao GD, Lin B, Huang XX, Song SJ. Modified lanostane-type triterpenoids with neuroprotective effects from the fungus Inonotus obliquus . Bioorg Chem. 2020; 105:104438. PMID: 33171406.20. Zou CX, Wang XB, Lv TM, Hou ZL, Lin B, Huang XX, Song SJ. Flavan derivative enantiomers and drimane sesquiterpene lactones from the Inonotus obliquus with neuroprotective effects. Bioorg Chem. 2020; 96:103588. PMID: 32032845.21. Zhu ZY, Liu XC, Fang XN, Sun HQ, Yang XY, Zhang YM. Structural characterization and anti-tumor activity of polysaccharide produced by Hirsutella sinensis . Int J Biol Macromol. 2016; 82:959–966. PMID: 26517958.22. Liu M, Liu Y, Cao MJ, Liu GM, Chen Q, Sun L, Chen H. Antibacterial activity and mechanisms of depolymerized fucoidans isolated from Laminaria japonica . Carbohydr Polym. 2017; 172:294–305. PMID: 28606538.23. Ahmad A, Ali T, Park HY, Badshah H, Rehman SU, Kim MO. Neuroprotective effect of fisetin against amyloid-beta-induced cognitive/synaptic dysfunction, neuroinflammation, and neurodegeneration in adult mice. Mol Neurobiol. 2017; 54:2269–2285. PMID: 26944285.24. Liu Y, Hettinger CL, Zhang D, Rezvani K, Wang X, Wang H. The proteasome function reporter GFPu accumulates in young brains of the APPswe/PS1dE9 Alzheimer’s disease mouse model. Cell Mol Neurobiol. 2014; 34:315–322. PMID: 24363091.25. Barrachina M, Castaño E, Dalfó E, Maes T, Buesa C, Ferrer I. Reduced ubiquitin C-terminal hydrolase-1 expression levels in dementia with Lewy bodies. Neurobiol Dis. 2006; 22:265–273. PMID: 16380264.26. Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004; 279:13256–13264. PMID: 14722078.27. Lonskaya I, Hebron ML, Desforges NM, Schachter JB, Moussa CE. Nilotinib-induced autophagic changes increase endogenous parkin level and ubiquitination, leading to amyloid clearance. J Mol Med (Berl). 2014; 92:373–386. PMID: 24337465.28. Pugazhenthi S, Qin L, Reddy PH. Common neurodegenerative pathways in obesity, diabetes, and Alzheimer’s disease. Biochim Biophys Acta BBAMol Basis Dis. 2017; 1863:1037–1045.29. Lu X, Chen H, Dong P, Fu L, Zhang X. Phytochemical characteristics and hypoglycaemic activity of fraction from mushroom Inonotus obliquus . J Sci Food Agric. 2010; 90:276–280. PMID: 20355042.30. Ma L, Chen H, Dong P, Lu X. Anti-inflammatory and anticancer activities of extracts and compounds from the mushroom Inonotus obliquus . Food Chem. 2013; 139:503–508. PMID: 23561137.31. Correia SC, Santos RX, Carvalho C, Cardoso S, Candeias E, Santos MS, Oliveira CR, Moreira PI. Insulin signaling, glucose metabolism and mitochondria: major players in Alzheimer’s disease and diabetes interrelation. Brain Res. 2012; 1441:64–78. PMID: 22290178.32. Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. BioEssays. 2008; 30:1172–1184. PMID: 18937370.33. Belfiore R, Rodin A, Ferreira E, Velazquez R, Branca C, Caccamo A, Oddo S. Temporal and regional progression of Alzheimer’s disease-like pathology in 3xTg-AD mice. Aging Cell. 2019; 18:e12873. PMID: 30488653.34. Watanabe T, Hikichi Y, Willuweit A, Shintani Y, Horiguchi T. FBL2 regulates amyloid precursor protein (APP) metabolism by promoting ubiquitination-dependent APP degradation and inhibition of APP endocytosis. J Neurosci. 2012; 32:3352–3365. PMID: 22399757.35. Borgegård T, Gustavsson S, Nilsson C, Parpal S, Klintenberg R, Berg AL, Rosqvist S, Serneels L, Svensson S, Olsson F, et al. Alzheimer’s disease: presenilin 2-sparing γ-secretase inhibition is a tolerable Aβ peptide-lowering strategy. J Neurosci. 2012; 32:17297–17305. PMID: 23197721.36. Garcia-Alloza M, Robbins EM, Zhang-Nunes SX, Purcell SM, Betensky RA, Raju S, Prada C, Greenberg SM, Bacskai BJ, Frosch MP. Characterization of amyloid deposition in the APPswe/PS1dE9 mouse model of Alzheimer disease. Neurobiol Dis. 2006; 24:516–524. PMID: 17029828.37. Ringman JM, Frautschy SA, Cole GM, Masterman DL, Cummings JL. A potential role of the curry spice curcumin in Alzheimer’s disease. Curr Alzheimer Res. 2005; 2:131–136. PMID: 15974909.38. Nalivaeva NN, Turner AJ. Targeting amyloid clearance in Alzheimer’s disease as a therapeutic strategy. Br J Pharmacol. 2019; 176:3447–3463. PMID: 30710367.39. Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, Yeh LA, Cuny GD, Stein RL, Lansbury PT Jr. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003; 10:837–846. PMID: 14522054.40. Gentier RJ, van Leeuwen FW. Misframed ubiquitin and impaired protein quality control: an early event in Alzheimer’s disease. Front Mol Neurosci. 2015; 8:47. PMID: 26388726.41. Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000; 75:436–439. PMID: 10854289.42. López Salon M, Morelli L, Castaño EM, Soto EF, Pasquini JM. Defective ubiquitination of cerebral proteins in Alzheimer’s disease. J Neurosci Res. 2000; 62:302–310. PMID: 11020223.43. Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006; 126:775–788. PMID: 16923396.44. Saido T, Leissring MA. Proteolytic degradation of amyloid β-protein. Cold Spring Harb Perspect Med. 2012; 2:a006379. PMID: 22675659.45. Layfield R, Lowe J, Bedford L. The ubiquitin-proteasome system and neurodegenerative disorders. Essays Biochem. 2005; 41:157–171. PMID: 16250904.46. Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson’s disease and other neurodegenerative diseases. Trends Cell Biol. 2004; 14:703–711. PMID: 15564047.47. Uddin MS, Mamun AA, Jakaria M, Thangapandiyan S, Ahmad J, Rahman MA, Mathew B, Abdel-Daim MM, Aleya L. Emerging promise of sulforaphane-mediated Nrf2 signaling cascade against neurological disorders. Sci Total Environ. 2020; 707:135624. PMID: 31784171.48. Upadhya SC, Hegde AN. Ubiquitin-proteasome pathway components as therapeutic targets for CNS maladies. Curr Pharm Des. 2005; 11:3807–3828. PMID: 16305513.49. Lonskaya I, Shekoyan AR, Hebron ML, Desforges N, Algarzae NK, Moussa CE. Diminished parkin solubility and co-localization with intraneuronal amyloid-β are associated with autophagic defects in Alzheimer’s disease. J Alzheimers Dis. 2013; 33:231–247. PMID: 22954671.50. Khandelwal PJ, Herman AM, Hoe HS, Rebeck GW, Moussa CE. Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Aβ in AD models. Hum Mol Genet. 2011; 20:2091–2102. PMID: 21378096.51. Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997; 89:115–126. PMID: 9094720.52. Solano RM, Casarejos MJ, Gómez A, Perucho J, de Yébenes JG, Mena MA. Parkin null cortical neuronal/glial cultures are resistant to amyloid-β1-42 toxicity: a role for autophagy? J Alzheimers Dis. 2012; 32:57–76. PMID: 22785397.53. Zhang M, Cai F, Zhang S, Zhang S, Song W. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer’s progression in vivo . Sci Rep. 2014; 4:7298. PMID: 25466238.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Animal Models of Alzheimer's Dementia
- Immunomodulatory Activity of the Water Extract from Medicinal Mushroom Inonotus obliquus
- Anticancer activity of subfractions containing pure compounds of Chaga mushroom (Inonotus obliquus) extract in human cancer cells and in Balbc/c mice bearing Sarcoma-180 cells
- Insight into the emerging and common experimental in‑vivo models of Alzheimer’s disease
- Assessment of Cognitive Phenotyping in Inbred, Genetically Modified Mice, and Transgenic Mouse Models of Alzheimer's Disease