Nutr Res Pract.
2023 Aug;17(4):597-615. 10.4162/nrp.2023.17.4.597.
Modulation of DNA methylation by one-carbon metabolism: a milestone for healthy aging
- Affiliations
-
- 1Chaum Life Center, CHA University School of Medicine, Seoul 06062, Korea
- 2Department of Nutrition, School of Public Health and Health Sciences, University of Massachusetts, Amherst, MA 01003, USA
- 3Unit of Internal Medicine B and ‘Epigenomics and Gene-Nutrient Interactions’ Laboratory, Department of Medicine, University of Verona School of Medicine, Policlinico “G.B. Rossi,” 37134 Verona, Italy
- KMID: 2545179
- DOI: http://doi.org/10.4162/nrp.2023.17.4.597
Abstract
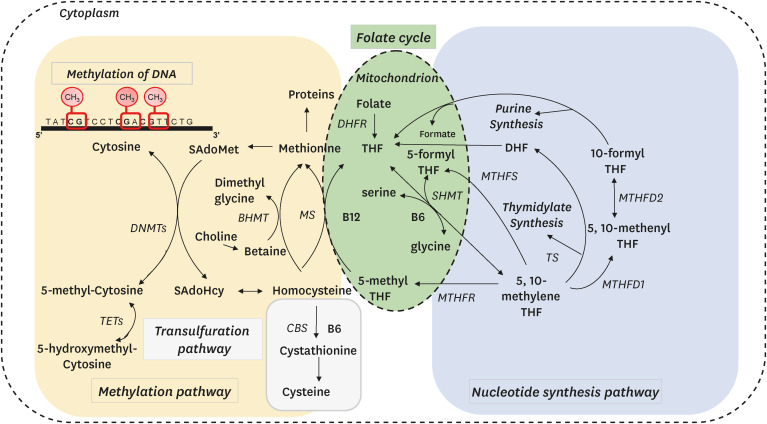
- Healthy aging can be defined as an extended lifespan and health span. Nutrition has been regarded as an important factor in healthy aging, because nutrients, bioactive food components, and diets have demonstrated beneficial effects on aging hallmarks such as oxidative stress, mitochondrial function, apoptosis and autophagy, genomic stability, and immune function. Nutrition also plays a role in epigenetic regulation of gene expression, and DNA methylation is the most extensively investigated epigenetic phenomenon in aging. Interestingly, age-associated DNA methylation can be modulated by one-carbon metabolism or inhibition of DNA methyltransferases. One-carbon metabolism ultimately controls the balance between the universal methyl donor S-adenosylmethionine and the methyltransferase inhibitor S-adenosylhomocysteine. Water-soluble B-vitamins such as folate, vitamin B6, and vitamin B12 serve as coenzymes for multiple steps in one-carbon metabolism, whereas methionine, choline, betaine, and serine act as methyl donors. Thus, these one-carbon nutrients can modify age-associated DNA methylation and subsequently alter the age-associated physiologic and pathologic processes. We cannot elude aging per se but we may at least change age-associated DNA methylation, which could mitigate age-associated diseases and disorders.
Keyword
Figure
Reference
-
1. Selvarani R, Mohammed S, Richardson A. Effect of rapamycin on aging and age-related diseases-past and future. Geroscience. 2021; 43:1135–1158. PMID: 33037985.
Article2. Bulterijs S, Hull RS, Björk VC, Roy AG. It is time to classify biological aging as a disease. Front Genet. 2015; 6:205. PMID: 26150825.
Article3. Fulop T, Larbi A, Dupuis G, Le Page A, Frost EH, Cohen AA, Witkowski JM, Franceschi C. Immunosenescence and inflamm-aging as two sides of the same coin: friends or foes? Front Immunol. 2018; 8:1960. PMID: 29375577.
Article4. Park LK, Friso S, Choi SW. Nutritional influences on epigenetics and age-related disease. Proc Nutr Soc. 2012; 71:75–83. PMID: 22051144.
Article5. Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013; 34:753–764. PMID: 22906839.
Article6. Johnstone SE, Gladyshev VN, Aryee MJ, Bernstein BE. Epigenetic clocks, aging, and cancer. Science. 2022; 378:1276–1277. PMID: 36548410.
Article7. Rahbari R, Wuster A, Lindsay SJ, Hardwick RJ, Alexandrov LB, Turki SA, Dominiczak A, Morris A, Porteous D, Smith B, et al. Timing, rates and spectra of human germline mutation. Nat Genet. 2016; 48:126–133. PMID: 26656846.
Article8. Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011; 3:503–518. PMID: 22022340.
Article9. Lan X, Field MS, Stover PJ. Cell cycle regulation of folate-mediated one-carbon metabolism. Wiley Interdiscip Rev Syst Biol Med. 2018; 10:e1426. PMID: 29889360.
Article10. Friso S, Udali S, De Santis D, Choi SW. One-carbon metabolism and epigenetics. Mol Aspects Med. 2017; 54:28–36. PMID: 27876555.
Article11. Marí M, de Gregorio E, de Dios C, Roca-Agujetas V, Cucarull B, Tutusaus A, Morales A, Colell A. Mitochondrial glutathione: recent insights and role in disease. Antioxidants (Basel). 2020; 9:909. PMID: 32987701.
Article12. Blom HJ, Smulders Y. Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects. J Inherit Metab Dis. 2011; 34:75–81. PMID: 20814827.
Article13. Mason JB. Biomarkers of nutrient exposure and status in one-carbon (methyl) metabolism. J Nutr. 2003; 133(Suppl 3):941S–947S. PMID: 12612180.
Article14. Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem. 2000; 275:29318–29323. PMID: 10884384.
Article15. Dahlhoff C, Worsch S, Sailer M, Hummel BA, Fiamoncini J, Uebel K, Obeid R, Scherling C, Geisel J, Bader BL, et al. Methyl-donor supplementation in obese mice prevents the progression of NAFLD, activates AMPK and decreases acyl-carnitine levels. Mol Metab. 2014; 3:565–580. PMID: 25061561.
Article16. Smith DE, Hornstra JM, Kok RM, Blom HJ, Smulders YM. Folic acid supplementation does not reduce intracellular homocysteine, and may disturb intracellular one-carbon metabolism. Clin Chem Lab Med. 2013; 51:1643–1650. PMID: 23740686.
Article17. Pizzolo F, Blom HJ, Choi SW, Girelli D, Guarini P, Martinelli N, Stanzial AM, Corrocher R, Olivieri O, Friso S. Folic acid effects on S-adenosylmethionine, S-adenosylhomocysteine, and DNA methylation in patients with intermediate hyperhomocysteinemia. J Am Coll Nutr. 2011; 30:11–18. PMID: 21697534.
Article18. Yao Y, Gao LJ, Zhou Y, Zhao JH, Lv Q, Dong JZ, Shang MS. Effect of advanced age on plasma homocysteine levels and its association with ischemic stroke in non-valvular atrial fibrillation. J Geriatr Cardiol. 2017; 14:743–749. PMID: 29581713.19. Kuo HK, Sorond FA, Chen JH, Hashmi A, Milberg WP, Lipsitz LA. The role of homocysteine in multisystem age-related problems: a systematic review. J Gerontol A Biol Sci Med Sci. 2005; 60:1190–1201. PMID: 16183962.
Article20. Ostrakhovitch EA, Tabibzadeh S. Homocysteine and age-associated disorders. Ageing Res Rev. 2019; 49:144–164. PMID: 30391754.
Article21. James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S-adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. J Nutr. 2002; 132:2361S–236S. PMID: 12163693.
Article22. Zhang DM, Ye JX, Mu JS, Cui XP. Efficacy of vitamin B supplementation on cognition in elderly patients with cognitive-related diseases. J Geriatr Psychiatry Neurol. 2017; 30:50–59. PMID: 28248558.
Article23. Li Y, Huang T, Zheng Y, Muka T, Troup J, Hu FB. Folic acid supplementation and the risk of cardiovascular diseases: a meta-analysis of randomized controlled trials. J Am Heart Assoc. 2016; 5:e003768. PMID: 27528407.
Article24. Annibal A, Tharyan RG, Schonewolff MF, Tam H, Latza C, Auler MM, Grönke S, Partridge L, Antebi A. Regulation of the one carbon folate cycle as a shared metabolic signature of longevity. Nat Commun. 2021; 12:3486. PMID: 34108489.
Article25. Pogribny IP, James SJ, Beland FA. Molecular alterations in hepatocarcinogenesis induced by dietary methyl deficiency. Mol Nutr Food Res. 2012; 56:116–125. PMID: 22095781.
Article26. Cheng TY, Makar KW, Neuhouser ML, Miller JW, Song X, Brown EC, Beresford SA, Zheng Y, Poole EM, Galbraith RL, et al. Folate-mediated one-carbon metabolism genes and interactions with nutritional factors on colorectal cancer risk: Women’s Health Initiative Observational Study. Cancer. 2015; 121:3684–3691. PMID: 26108676.
Article27. Murray B, Barbier-Torres L, Fan W, Mato JM, Lu SC. Methionine adenosyltransferases in liver cancer. World J Gastroenterol. 2019; 25:4300–4319. PMID: 31496615.
Article28. Rushworth D, Mathews A, Alpert A, Cooper LJ. Dihydrofolate reductase and thymidylate synthase transgenes resistant to methotrexate interact to permit novel transgene regulation. J Biol Chem. 2015; 290:22970–22976. PMID: 26242737.
Article29. Showalter SL, Showalter TN, Witkiewicz A, Havens R, Kennedy EP, Hucl T, Kern SE, Yeo CJ, Brody JR. Evaluating the drug-target relationship between thymidylate synthase expression and tumor response to 5-fluorouracil. Is it time to move forward? Cancer Biol Ther. 2008; 7:986–994. PMID: 18443433.
Article30. Raghubeer S, Matsha TE. Methylenetetrahydrofolate (MTHFR), the one-carbon cycle, and cardiovascular risks. Nutrients. 2021; 13:4562. PMID: 34960114.
Article31. Nieraad H, de Bruin N, Arne O, Hofmann MC, Schmidt M, Saito T, Saido TC, Gurke R, Schmidt D, Till U, et al. Impact of hyperhomocysteinemia and different dietary interventions on cognitive performance in a knock-in mouse model for Alzheimer’s disease. Nutrients. 2020; 12:4562.
Article32. Zeng J, Gu Y, Fu H, Liu C, Zou Y, Chang H. Association between one-carbon metabolism-related vitamins and risk of breast cancer: a systematic review and meta-analysis of prospective studies. Clin Breast Cancer. 2020; 20:e469–e480. PMID: 32241696.
Article33. Bønaa KH, Njølstad I, Ueland PM, Schirmer H, Tverdal A, Steigen T, Wang H, Nordrehaug JE, Arnesen E, Rasmussen K, et al. Homocysteine lowering and cardiovascular events after acute myocardial infarction. N Engl J Med. 2006; 354:1578–1588. PMID: 16531614.
Article34. Lonn E, Yusuf S, Arnold MJ, Sheridan P, Pogue J, Micks M, McQueen MJ, Probstfield J, Fodor G, Held C, et al. Homocysteine lowering with folic acid and B vitamins in vascular disease. N Engl J Med. 2006; 354:1567–1577. PMID: 16531613.
Article35. Kwok T, Wu Y, Lee J, Lee R, Yung CY, Choi G, Lee V, Harrison J, Lam L, Mok V. A randomized placebo-controlled trial of using B vitamins to prevent cognitive decline in older mild cognitive impairment patients. Clin Nutr. 2020; 39:2399–2405. PMID: 31787369.
Article36. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009; 324:930–935. PMID: 19372391.
Article37. Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011; 25:679–684. PMID: 21460036.
Article38. Hu YC, Hu HC, Lin S, Chen XM. The role of DNA hydroxymethylation in the regulation of atherosclerosis. Yi Chuan. 2020; 42:632–640. PMID: 32694103.39. Li Y, Liu Y, Strickland FM, Richardson B. Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity and acute coronary syndromes. Exp Gerontol. 2010; 45:312–322. PMID: 20035856.
Article40. Arand J, Chiang HR, Martin D, Snyder MP, Sage J, Reijo Pera RA, Wossidlo M. Tet enzymes are essential for early embryogenesis and completion of embryonic genome activation. EMBO Rep. 2022; 23:e53968. PMID: 34866320.
Article41. Johnson ND, Huang L, Li R, Li Y, Yang Y, Kim HR, Grant C, Wu H, Whitsel EA, Kiel DP, et al. Age-related DNA hydroxymethylation is enriched for gene expression and immune system processes in human peripheral blood. Epigenetics. 2020; 15:294–306. PMID: 31506003.
Article42. Tammen SA, Dolnikowski GG, Ausman LM, Liu Z, Kim KC, Friso S, Choi SW. Aging alters hepatic DNA hydroxymethylation, as measured by liquid chromatography/mass spectrometry. J Cancer Prev. 2014; 19:301–308. PMID: 25574465.
Article43. Truong TP, Sakata-Yanagimoto M, Yamada M, Nagae G, Enami T, Nakamoto-Matsubara R, Aburatani H, Chiba S. Age-dependent decrease of DNA hydroxymethylation in human T cells. J Clin Exp Hematop. 2015; 55:1–6. PMID: 26105999.
Article44. Tsurumi A, Li WX. Global heterochromatin loss: a unifying theory of aging? Epigenetics. 2012; 7:680–688. PMID: 22647267.45. Lee JH, Kim EW, Croteau DL, Bohr VA. Heterochromatin: an epigenetic point of view in aging. Exp Mol Med. 2020; 52:1466–1474. PMID: 32887933.
Article46. Harrison A, Parle-McDermott A. DNA methylation: a timeline of methods and applications. Front Genet. 2011; 2:74. PMID: 22303369.
Article47. Ashapkin VV, Kutueva LI, Vanyushin BF. Aging as an epigenetic phenomenon. Curr Genomics. 2017; 18:385–407. PMID: 29081695.
Article48. Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012; 109:10522–10527. PMID: 22689993.
Article49. Sun D, Yi SV. Impacts of chromatin states and long-range genomic segments on aging and DNA methylation. PLoS One. 2015; 10:e0128517. PMID: 26091484.
Article50. Corso-Díaz X, Gentry J, Rebernick R, Jaeger C, Brooks MJ, van Asten F, Kooragayala K, Gieser L, Nellissery J, Covian R, et al. Genome-wide profiling identifies DNA methylation signatures of aging in rod photoreceptors associated with alterations in energy metabolism. Cell Reports. 2020; 31:107525. PMID: 32320661.
Article51. Zhao M, Qin J, Yin H, Tan Y, Liao W, Liu Q, Luo S, He M, Liang G, Shi Y, et al. Distinct epigenomes in CD4+ T cells of newborns, middle-ages and centenarians. Sci Rep. 2016; 6:38411. PMID: 27917918.52. Johansson A, Enroth S, Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS One. 2013; 8:e67378. PMID: 23826282.
Article53. Unnikrishnan A, Freeman WM, Jackson J, Wren JD, Porter H, Richardson A. The role of DNA methylation in epigenetics of aging. Pharmacol Ther. 2019; 195:172–185. PMID: 30419258.
Article54. Bell CG, Lowe R, Adams PD, Baccarelli AA, Beck S, Bell JT, Christensen BC, Gladyshev VN, Heijmans BT, Horvath S, et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 2019; 20:249. PMID: 31767039.
Article55. Saul D, Kosinsky RL. Epigenetics of aging and aging-associated diseases. Int J Mol Sci. 2021; 22:401. PMID: 33401659.
Article56. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013; 14:R115. PMID: 24138928.
Article57. Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013; 49:359–367. PMID: 23177740.
Article58. Dhingra R, Kwee LC, Diaz-Sanchez D, Devlin RB, Cascio W, Hauser ER, Gregory S, Shah S, Kraus WE, Olden K, et al. Evaluating DNA methylation age on the Illumina MethylationEPIC Bead Chip. PLoS One. 2019; 14:e0207834. PMID: 31002714.
Article59. Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine. 2017; 21:29–36. PMID: 28396265.
Article60. Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018; 10:573–591. PMID: 29676998.
Article61. Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019; 11:303–327. PMID: 30669119.
Article62. Nwanaji-Enwerem JC, Colicino E, Gao X, Wang C, Vokonas P, Boyer EW, Baccarelli AA, Schwartz J. Associations of plasma folate and vitamin B6 with blood DNA methylation age: an analysis of one-carbon metabolites in the VA Normative Aging Study. J Gerontol A Biol Sci Med Sci. 2021; 76:760–769. PMID: 33027507.
Article63. Fitzgerald KN, Hodges R, Hanes D, Stack E, Cheishvili D, Szyf M, Henkel J, Twedt MW, Giannopoulou D, Herdell J, et al. Potential reversal of epigenetic age using a diet and lifestyle intervention: a pilot randomized clinical trial. Aging (Albany NY). 2021; 13:9419–9432. PMID: 33844651.
Article64. Kim KC, Friso S, Choi SW. DNA methylation, an epigenetic mechanism connecting folate to healthy embryonic development and aging. J Nutr Biochem. 2009; 20:917–926. PMID: 19733471.
Article65. Lillycrop KA, Hoile SP, Grenfell L, Burdge GC. DNA methylation, ageing and the influence of early life nutrition. Proc Nutr Soc. 2014; 73:413–421. PMID: 25027290.
Article66. Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014; 28:812–828. PMID: 24736841.
Article67. Fernández-Arroyo S, Cuyàs E, Bosch-Barrera J, Alarcón T, Joven J, Menendez JA. Activation of the methylation cycle in cells reprogrammed into a stem cell-like state. Oncoscience. 2016; 2:958–967. PMID: 26909364.
Article68. Van Winkle LJ, Ryznar R. One-carbon metabolism regulates embryonic stem cell fate through epigenetic DNA and histone modifications: implications for transgenerational metabolic disorders in adults. Front Cell Dev Biol. 2019; 7:300. PMID: 31824950.
Article69. Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC. Erice Imprinting Group. Genomic imprinting and physiological processes in mammals. Cell. 2019; 176:952–965. PMID: 30794780.
Article70. Dunford AR, Sangster JM. Maternal and paternal periconceptional nutrition as an indicator of offspring metabolic syndrome risk in later life through epigenetic imprinting: A systematic review. Diabetes Metab Syndr. 2017; 11(Suppl 2):S655–S662. PMID: 28533070.71. Chamberlain JA, Dugué PA, Bassett JK, Hodge AM, Brinkman MT, Joo JE, Jung CH, Makalic E, Schmidt DF, Hopper JL, et al. Dietary intake of one-carbon metabolism nutrients and DNA methylation in peripheral blood. Am J Clin Nutr. 2018; 108:611–621. PMID: 30101351.
Article72. Ozias MK, Schalinske KL. All-trans-retinoic acid rapidly induces glycine N-methyltransferase in a dose-dependent manner and reduces circulating methionine and homocysteine levels in rats. J Nutr. 2003; 133:4090–4094. PMID: 14652353.
Article73. Speckmann B, Schulz S, Hiller F, Hesse D, Schumacher F, Kleuser B, Geisel J, Obeid R, Grune T, Kipp AP. Selenium increases hepatic DNA methylation and modulates one-carbon metabolism in the liver of mice. J Nutr Biochem. 2017; 48:112–119. PMID: 28810182.
Article74. Wischhusen P, Saito T, Heraud C, Kaushik SJ, Fauconneau B, Antony Jesu Prabhu P, Fontagné-Dicharry S, Skjærven KH. Parental selenium nutrition affects the one-carbon metabolism and the hepatic DNA methylation pattern of rainbow trout (Oncorhynchus mykiss) in the progeny. Life (Basel). 2020; 10:121. PMID: 32722369.
Article75. Mierziak J, Kostyn K, Boba A, Czemplik M, Kulma A, Wojtasik W. Influence of the bioactive diet components on the gene expression regulation. Nutrients. 2021; 13:3673. PMID: 34835928.76. Hassan FU, Rehman MS, Khan MS, Ali MA, Javed A, Nawaz A, Yang C. Curcumin as an alternative epigenetic modulator: mechanism of action and potential effects. Front Genet. 2019; 10:514. PMID: 31214247.
Article77. Farhan M, Ullah MF, Faisal M, Farooqi AA, Sabitaliyevich UY, Biersack B, Ahmad A. Differential methylation and acetylation as the epigenetic basis of resveratrol’s anticancer activity. Medicines (Basel). 2019; 6:24. PMID: 30781847.
Article78. Sae-Lee C, Corsi S, Barrow TM, Kuhnle GG, Bollati V, Mathers JC, Byun HM. Dietary intervention modifies DNA methylation age assessed by the epigenetic clock. Mol Nutr Food Res. 2018; 62:e1800092. PMID: 30350398.
Article79. Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002; 99:5606–5611. PMID: 11929966.
Article80. Mohammad K, Titorenko VI. Caloric restriction creates a metabolic pattern of chronological aging delay that in budding yeast differs from the metabolic design established by two other geroprotectors. Oncotarget. 2021; 12:608–625. PMID: 33868583.81. Kim CH, Lee EK, Choi YJ, An HJ, Jeong HO, Park D, Kim BC, Yu BP, Bhak J, Chung HY. Short-term calorie restriction ameliorates genomewide, age-related alterations in DNA methylation. Aging Cell. 2016; 15:1074–1081. PMID: 27561685.82. Sziráki A, Tyshkovskiy A, Gladyshev VN. Global remodeling of the mouse DNA methylome during aging and in response to calorie restriction. Aging Cell. 2018; 17:e12738. PMID: 29575528.
Article83. Lu Y, Brommer B, Tian X, Krishnan A, Meer M, Wang C, Vera DL, Zeng Q, Yu D, Bonkowski MS, et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature. 2020; 588:124–129. PMID: 33268865.84. Green DR. Polyamines and aging: a CLEAR connection? Mol Cell. 2019; 76:5–7. PMID: 31585103.85. Aissa AF, Amaral CL, Venancio VP, Machado CD, Hernandes LC, Santos PW, Curi R, Bianchi ML, Antunes LM. Methionine-supplemented diet affects the expression of cardiovascular disease-related genes and increases inflammatory cytokines in mice heart and liver. J Toxicol Environ Health A. 2017; 80:1116–1128. PMID: 28880739.86. Virtanen JK, Voutilainen S, Rissanen TH, Happonen P, Mursu J, Laukkanen JA, Poulsen H, Lakka TA, Salonen JT. High dietary methionine intake increases the risk of acute coronary events in middle-aged men. Nutr Metab Cardiovasc Dis. 2006; 16:113–120. PMID: 16487911.87. Ables GP, Hens JR, Nichenametla SN. Methionine restriction beyond life-span extension. Ann N Y Acad Sci. 2016; 1363:68–79. PMID: 26916321.88. Kitada M, Ogura Y, Monno I, Xu J, Koya D. Effect of methionine restriction on aging: its relationship to oxidative stress. Biomedicines. 2021; 9:130. PMID: 33572965.89. Sanderson SM, Gao X, Dai Z, Locasale JW. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat Rev Cancer. 2019; 19:625–637. PMID: 31515518.90. Wanders D, Hobson K, Ji X. Methionine restriction and cancer biology. Nutrients. 2020; 12:684. PMID: 32138282.
Article91. Lauinger L, Kaiser P. Sensing and signaling of methionine metabolism. Metabolites. 2021; 11:83. PMID: 33572567.
Article92. Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP, Sabatini DM. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 2017; 358:813–818. PMID: 29123071.93. Mattocks DA, Mentch SJ, Shneyder J, Ables GP, Sun D, Richie JP Jr, Locasale JW, Nichenametla SN. Short term methionine restriction increases hepatic global DNA methylation in adult but not young male C57BL/6J mice. Exp Gerontol. 2017; 88:1–8. PMID: 27940170.94. Tamanna N, Mayengbam S, House JD, Treberg JR. Methionine restriction leads to hyperhomocysteinemia and alters hepatic H2S production capacity in Fischer-344 rats. Mech Ageing Dev. 2018; 176:9–18. PMID: 30367932.95. Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. S-adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc Natl Acad Sci U S A. 2006; 103:6489–6494. PMID: 16614071.96. Wong M. Mammalian target of rapamycin (mTOR) pathways in neurological diseases. Biomed J. 2013; 36:40–50. PMID: 23644232.
Article97. Ogawa T, Masumura K, Kohara Y, Kanai M, Soga T, Ohya Y, Blackwell TK, Mizunuma M. S-adenosyl-L-homocysteine extends lifespan through methionine restriction effects. Aging Cell. 2022; 21:e13604. PMID: 35388610.98. Reddy VS, Trinath J, Reddy GB. Implication of homocysteine in protein quality control processes. Biochimie. 2019; 165:19–31. PMID: 31269461.
Article99. Ganguly P, Alam SF. Role of homocysteine in the development of cardiovascular disease. Nutr J. 2015; 14:6. PMID: 25577237.100. Luzzi S, Cherubini V, Falsetti L, Viticchi G, Silvestrini M, Toraldo A. Homocysteine, cognitive functions, and degenerative dementias: state of the art. Biomedicines. 2022; 10:2741. PMID: 36359260.101. Burgos-Barragan G, Wit N, Meiser J, Dingler FA, Pietzke M, Mulderrig L, Pontel LB, Rosado IV, Brewer TF, Cordell RL, et al. Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature. 2017; 548:549–554. PMID: 28813411.102. Schug ZT. Formaldehyde detoxification creates a new wheel for the folate-driven one-carbon “bi”-cycle. Biochemistry. 2018; 57:889–890. PMID: 29368500.
Article103. He H, Noor E, Ramos-Parra PA, Garcia-Valencia LE, Patterson JA, Diaz de la Garza RI, Hanson AD, Bar-Even A. In vivo rate of formaldehyde condensation with tetrahydrofolate. Metabolites. 2020; 10:65–60. PMID: 32059429.
Article104. Chen X, Chothia SY, Basran J, Hopkinson RJ. Formaldehyde regulates tetrahydrofolate stability and thymidylate synthase catalysis. Chem Commun (Camb). 2021; 57:5778–5781. PMID: 33997872.105. Morellato AE, Umansky C, Pontel LB. The toxic side of one-carbon metabolism and epigenetics. Redox Biol. 2021; 40:101850. PMID: 33418141.106. Brosnan ME, Brosnan JT. Formate: the neglected member of one-carbon metabolism. Annu Rev Nutr. 2016; 36:369–388. PMID: 27431368.107. Yang JH, Hayano M, Griffin PT, Amorim JA, Bonkowski MS, Apostolides JK, Salfati EL, Blanchette M, Munding EM, Bhakta M, et al. Loss of epigenetic information as a cause of mammalian aging. Cell. 2023; 186:305–326.e27. PMID: 36638792.