Cancer Res Treat.
2020 Jan;52(1):98-108. 10.4143/crt.2019.195.
High-Throughput Multiplex Immunohistochemical Imaging of the Tumor and Its Microenvironment
- Affiliations
-
- 1Department of Pathology, Seoul National University Hospital, Seoul, Korea
- 2Department of Pathology, Seoul National University Bundang Hospital, Seongnam, Korea
- 3Cancer Research Institute, Seoul National University, Seoul, Korea
- 4Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2501205
- DOI: http://doi.org/10.4143/crt.2019.195
Abstract
- Purpose
The aim of this study was to develop a formalin-fixed paraffin-embedded (FFPE) tissue based multiplex immunochemistry (mIHC) method for high-throughput comprehensive tissue imaging and demonstrate its feasibility, validity, and usefulness.
Materials and Methods
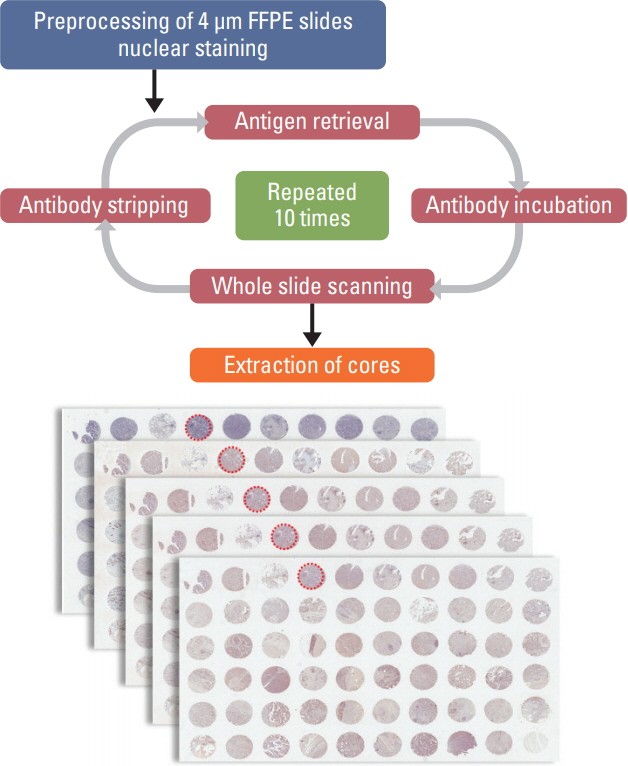
The mIHC protocol was developed and tested on tissue microarray slides made from archived gastric cancer (GC) tissue samples. On a single FFPE slide, cyclic immunochemistry for multiple markers of immune cells and cytokeratin for tumor cells was performed; hematoxylin staining was used for demarcation of nuclei. Whole slides were digitally scanned after each cycle. For interpretation of mIHC results, we performed computer-assisted image analysis using publicly available software.
Results
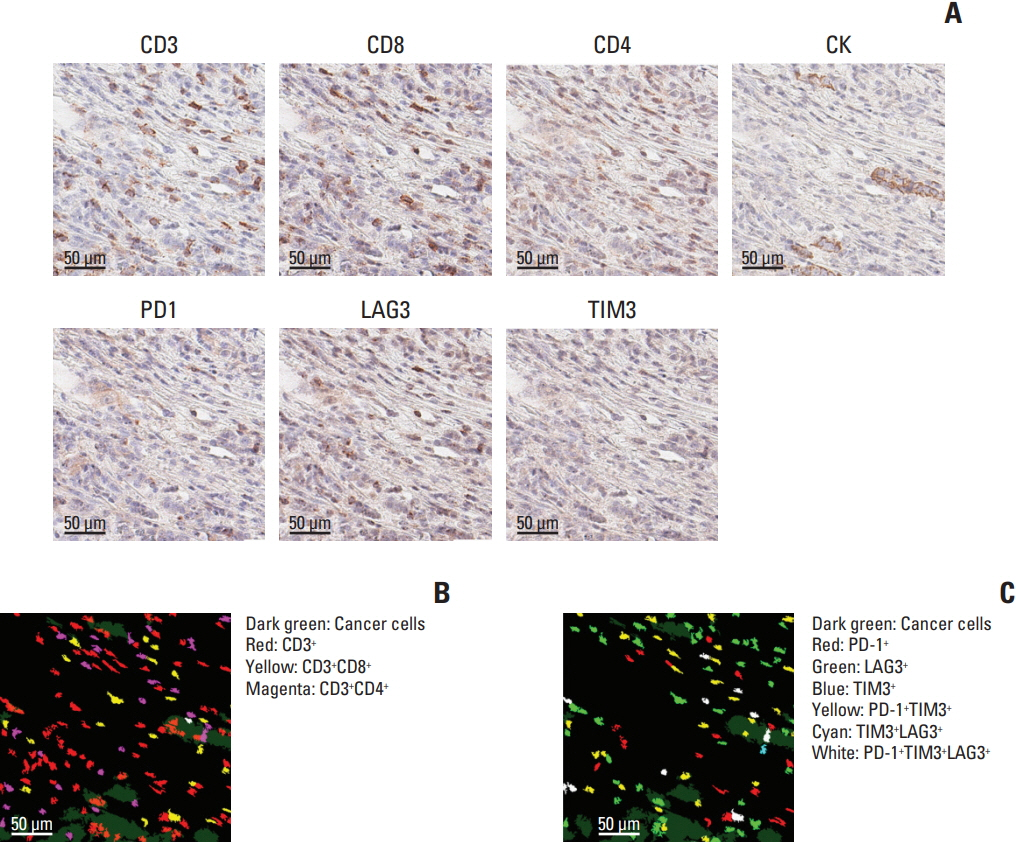
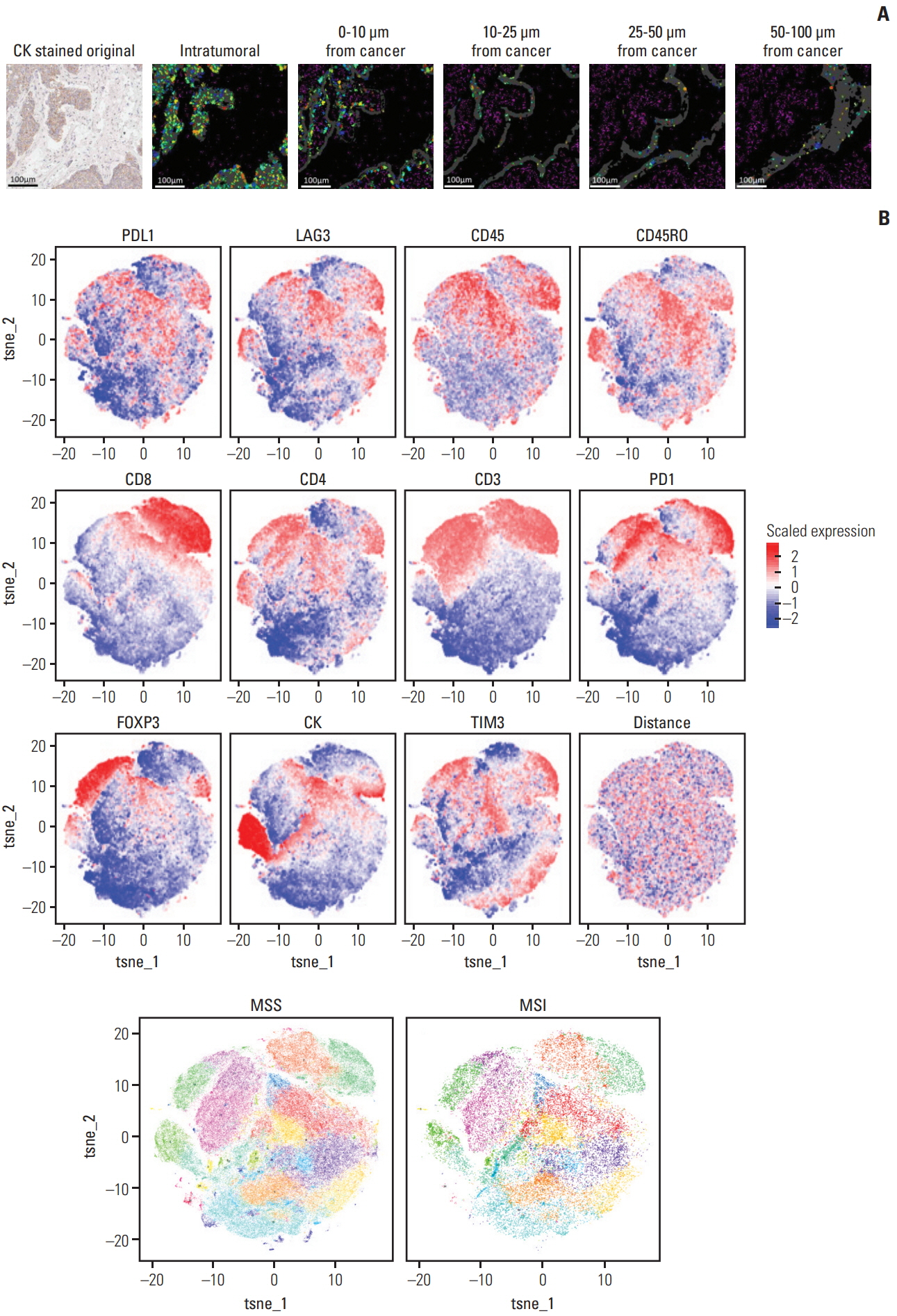
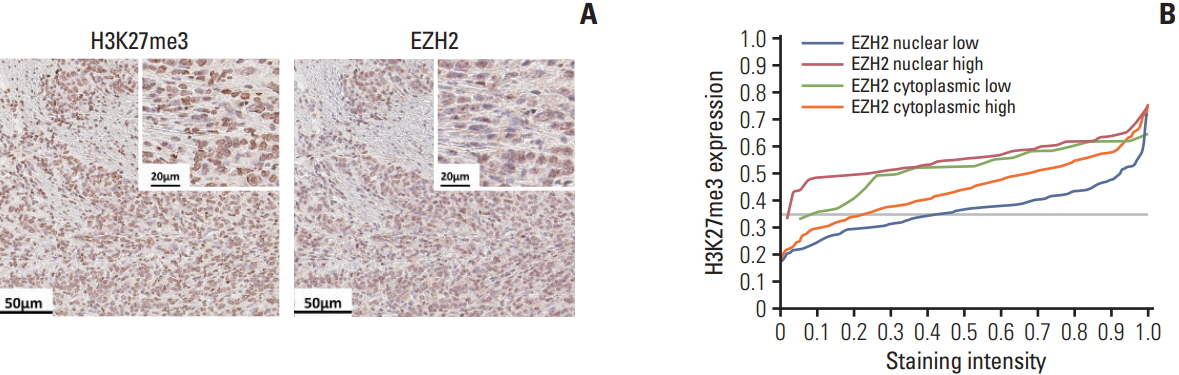
Using mIHC, we were able to characterize the tumor microenvironment (TME) of GCs with accurate visualization of various immune cells harboring complex immunophenotypes. Spatial information regarding intratumoral and peritumoral TME could be demonstrated by digital segmentation of image guided by cytokeratin staining results. We further extended the application of mIHC by showing that subcellular localization of molecules can be achieved by image analysis of mIHC results.
Conclusion
We developed a robust method for high-throughput multiplex imaging of FFPE tissue slides. The feasibility and adaptability of mIHC suggest that it is an efficient method for in situ single-cell characterization and analysis.
Keyword
Figure
Reference
-
References
1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–74.
Article2. Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010; 363:711–23.
Article3. Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015; 372:311–9.
Article4. Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015; 372:2018–28.
Article5. Mlecnik B, Van den Eynde M, Bindea G, Church SE, Vasaturo A, Fredriksen T, et al. Comprehensive intrametastatic immune quantification and major impact of immunoscore on survival. J Natl Cancer Inst. 2018; 110:97–108.
Article6. Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017; 19:203–17.
Article7. Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014; 11:417–22.
Article8. Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018; 175:984–97.
Article9. Lee KS, Nam SK, Koh J, Kim DW, Kang SB, Choe G, et al. Stromal expression of microRNA-21 in advanced colorectal cancer patients with distant metastases. J Pathol Transl Med. 2016; 50:270–7.
Article10. Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, et al. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics. 2008; 9:482.
Article11. Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013; 31:545–52.
Article12. Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004; 96:261–8.
Article13. Natsume A, Ito M, Katsushima K, Ohka F, Hatanaka A, Shinjo K, et al. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in glioblastoma. Cancer Res. 2013; 73:4559–70.
Article14. Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005; 121:425–36.
Article15. Bataille F, Troppmann S, Klebl F, Rogler G, Stoelcker B, Hofstadter F, et al. Multiparameter immunofluorescence on paraffin-embedded tissue sections. Appl Immunohistochem Mol Morphol. 2006; 14:225–8.
Article16. Viegas MS, Martins TC, Seco F, do Carmo A. An improved and cost-effective methodology for the reduction of autofluo rescence in direct immunofluorescence studies on formalinfixed paraffin-embedded tissues. Eur J Histochem. 2007; 51:59–66.17. Robertson D, Savage K, Reis-Filho JS, Isacke CM. Multiple immunofluorescence labelling of formalin-fixed paraffinembedded (FFPE) tissue. BMC Cell Biol. 2008; 9:13.
Article18. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014; 513:202–9.19. Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003; 100:10393–8.
Article20. Qiu P, Simonds EF, Bendall SC, Gibbs KD Jr, Bruggner RV, Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011; 29:886–91.
Article21. Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A. 2014; 111:E2770–7.
Article22. Taylor CR, Levenson RM. Quantification of immunohistochemistry: issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006; 49:411–24.23. Mengel M, von Wasielewski R, Wiese B, Rudiger T, Muller-Hermelink HK, Kreipe H. Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J Pathol. 2002; 198:292–9.
Article24. Howat WJ, Lewis A, Jones P, Kampf C, Ponten F, van der Loos CM, et al. Antibody validation of immunohistochemistry for biomarker discovery: recommendations of a consortium of academic and pharmaceutical based histopathology researchers. Methods. 2014; 70:34–8.
Article25. O'Hurley G, Sjostedt E, Rahman A, Li B, Kampf C, Ponten F, et al. Garbage in, garbage out: a critical evaluation of strategies used for validation of immunohistochemical biomarkers. Mol Oncol. 2014; 8:783–98.26. Remark R, Merghoub T, Grabe N, Litjens G, Damotte D, Wolchok JD, et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol. 2016; 1:aaf6925.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Tissue-Engineered 3D In Vitro Disease Models for HighThroughput Drug Screening
- Comparative Assessment of Diagnostic Performance of Cytochrome Oxidase Multiplex PCR and 18S rRNA Nested PCR
- Crosstalk between tumor-associated macrophages and neighboring cells in hepatocellular carcinoma
- Imaging in Tumor Immunology
- Exosomal Communication Between the Tumor Microenvironment and Innate Immunity and Its Therapeutic Application