Korean J Radiol.
2005 Dec;6(4):221-228. 10.3348/kjr.2005.6.4.221.
Mammographic Mass Detection Using a Mass Template
- Affiliations
-
- 1Istanbul Commerce University, RagIp, Gumuspala Cad. No: 84 Eminonu 34378, Istanbul, Turkey. serhat@iticu.edu.tr
- 2Marmara University, Goztepe, 81040, Istanbul, Turkey.
- KMID: 1777279
- DOI: http://doi.org/10.3348/kjr.2005.6.4.221
Abstract
OBJECTIVE
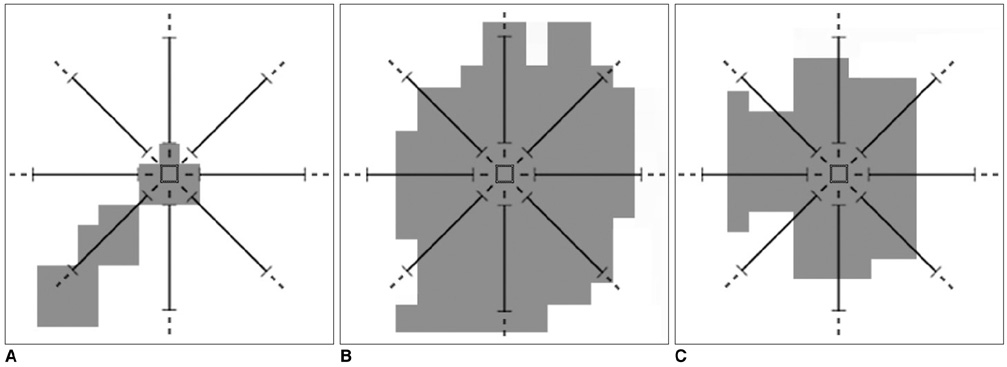
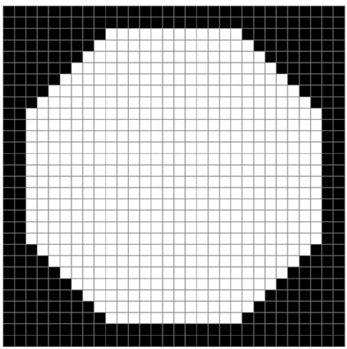
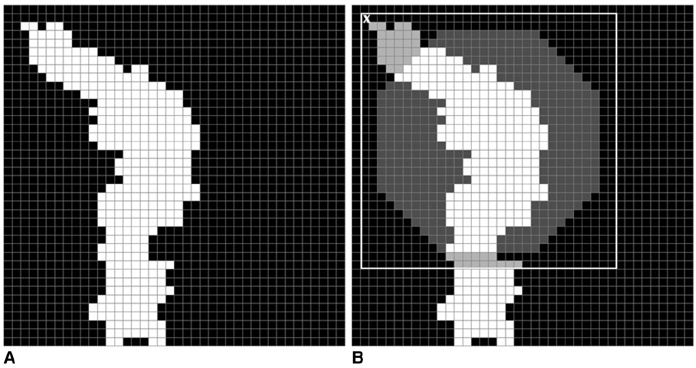
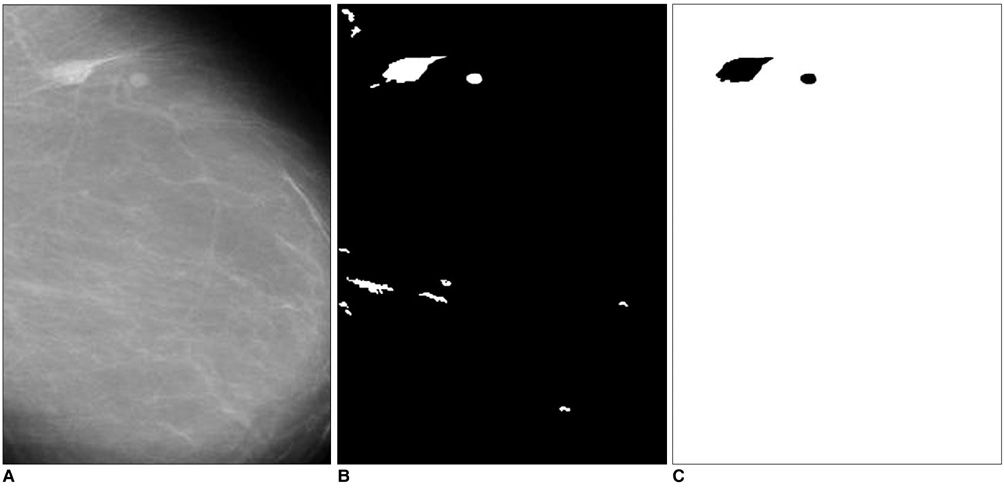
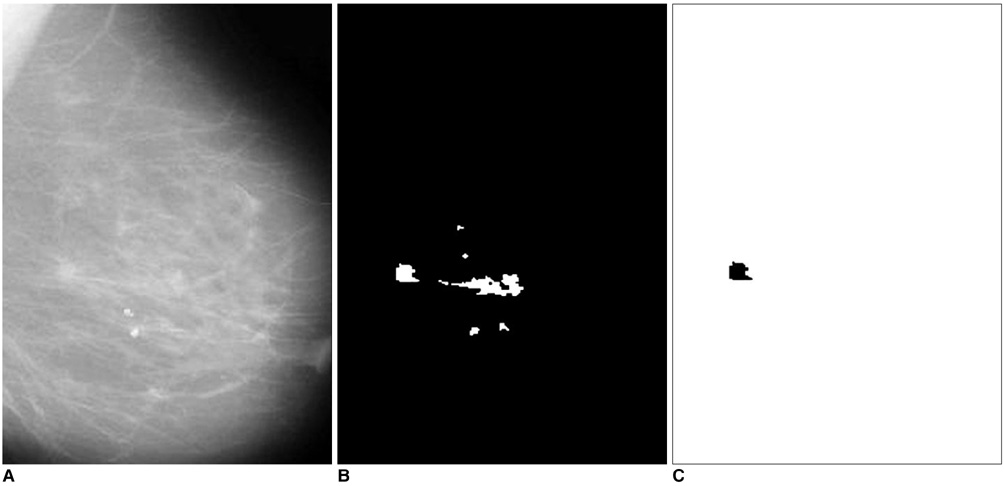
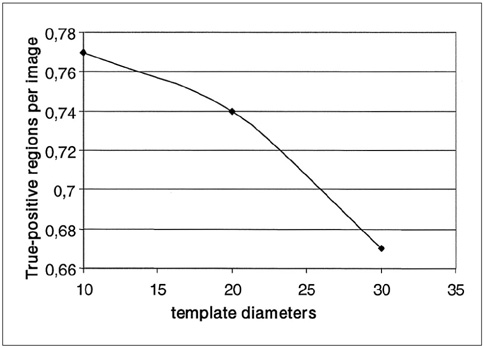
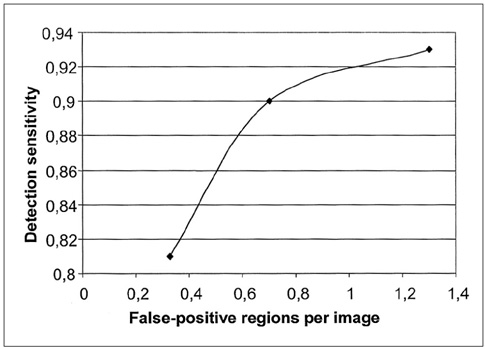
The purpose of this study was to develop a new method for automated mass detection in digital mammographic images using templates. MATERIALS AND METHODS: Masses were detected using a two steps process. First, the pixels in the mammogram images were scanned in 8 directions, and regions of interest (ROI) were identified using various thresholds. Then, a mass template was used to categorize the ROI as true masses or non-masses based on their morphologies. Each pixel of a ROI was scanned with a mass template to determine whether there was a shape (part of a ROI) similar to the mass in the template. The similarity was controlled using two thresholds. If a shape was detected, then the coordinates of the shape were recorded as part of a true mass. To test the system's efficiency, we applied this process to 52 mammogram images from the Mammographic Image Analysis Society (MIAS) database. RESULTS: Three hundred and thirty-two ROI were identified using the ROI specification methods. These ROI were classified using three templates whose diameters were 10, 20 and 30 pixels. The results of this experiment showed that using the templates with these diameters achieved sensitivities of 93%, 90% and 81% with 1.3, 0.7 and 0.33 false positives per image respectively. CONCLUSION: These results indicate that the detection performance of this template based algorithm is satisfactory, and may improve the performance of computer-aided analysis of mammographic images and early diagnosis of mammographic masses.
MeSH Terms
Figure
Cited by 1 articles
-
An Engineering View on Megatrends in Radiology: Digitization to Quantitative Tools of Medicine
Namkug Kim, Jaesoon Choi, Jaeyoun Yi, Seungwook Choi, Seyoun Park, Yongjun Chang, Joon Beom Seo
Korean J Radiol. 2013;14(2):139-153. doi: 10.3348/kjr.2013.14.2.139.
Reference
-
1. Baines CJ, McFarlane DV, Miller AB. The role of the reference radiologist: Estimates of interobserver agreement and potential delay in cancer detection in the national screening study. Invest Radiol. 1990. 25:971–976.2. Wallis M, Walsh M, Lee J. A review of false negative mammography in a symptomatic population. Clin Radiol. 1991. 44:13–15.3. Bird RE. Professional quality assurance for mammography screening programs. Radiology. 1990. 177:587.4. Brenner RJ. Medicolegal aspects of breast imaging: variable standards of care relating to different types of practice. AJR Am J Roentgenol. 1991. 156:719–723.5. Thurfjell EL, Lernevall KA, Taube AA. Benefit of independent double reading in a population-based mammography screening program. Radiology. 1994. 191:241–244.6. Vyborny CJ, Giger ML. Computer vision and artificial intelligence in mammography. AJR Am J Roentgenol. 1994. 162:699–708.7. Suckling J, Parker J, Dance D, Astley S, Hutt I, Boggis C, et al. The mammographic images analysis society digital mammogram database. Exerpta Med. 1994. 1069:375–378.8. Castellino RA, Roehrig J, Zhang W. Improved computer aided detection (CAD) algorithms for screening mammography. Radiology. 2000. 217:400.9. Malich A, Marx C, Facius M, Boehm T, Fleck M, Kaiser WA. Tumor detection rate of a new commercially available computer-aided detection system. Eur Radiol. 2001. 11:2454–2459.10. Karssemeijer N, Otten JD, Verbeek AL, Groenewoud JH, de Koning HJ, Hendriks JH, et al. Computer-aided detection versus independent double reading of masses on mammograms. Radiology. 2003. 227:192–200.11. Brzakovic D, Neskovic M. Mammogram screening using multiresolution-based image segmentation. Intern J Pattern Recognit Artif Intell. 1993. 7:1437–1460.12. Petrick N, Chan HP, Wei D. An adaptive density-weighted contrast enhancement filter for mammograhpic breast mass detection. IEEE Trans Med Imaging. 1996. 15:59–67.13. Chan H, Wei D, Helvie N, Sahiner B, Adler D, Goodsitt M, et al. Computer-aided classification of mammographic masses and normal tissue: linear discriminant analysis in texture feature space. Phys Med Biol. 1995. 40:857–876.14. Yin FF, Giger ML, Vyborny CJ, Doi K, Schmidt RA. Comparison of bilateral-subtraction and single-image processing techniques in the computerized detection of mammographic masses. Invest Radiol. 1993. 28:473–481.15. Kegelmeyer WP, Pruneda JM, Bourland PD, Hillis A, Riggs MW, Nipper ML. Computer-aided mammographic screening for spiculated lesions. Radiology. 1994. 191:331–337.16. Petrick N, Chan HP, Wei D, Sahiner B, Helvie MA, Adler DD. Automated detection of breast masses on mammograms using adaptive contrast enhancement and texture classification. Med Phy. 1996. 23:1685–1696.17. Singh S, Bovis K. An evaluation of contrast enhancement techniques for mammographic breast masses. IEEE Trans Inf Technol Biomed. 2005. 9:109–119.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Analysis of X-ray Mammographic Findings of Breast Carcinoma according to Histopathologic Classification
- Paget's Disease of the Breast: Significance of Mammographic Findings
- Ultrasonogphy in Paget's Disease of the Breast: Comparison with Mammographic Findings
- The Biologic Significance of Mammographic Calcification
- Nonpalpable Breast Cancer: Mammographic and Clinical findings